Abstract
Bacillus subtilis is capable of degrading fructosamines. The phosphorylation and the cleavage of the resulting fructosamine 6-phosphates is catalyzed by the frlD and frlB gene products, respectively. This study addresses the physiological importance of the frlBONMD genes (formerly yurPONML), revealing the necessity of their expression for growth on fructosamines and focusing on the complex regulation of the corresponding transcription unit. In addition to the known regulation by the global transcriptional regulator CodY, the frl genes are repressed by the convergently transcribed FrlR (formerly YurK). The latter causes repression during growth on substrates other than fructosamines. Additionally, we identified in the first intergenic region of the operon an FrlR binding site which is centrally located within a 38-bp perfect palindromic sequence. There is genetic evidence that this sequence, in combination with FrlR, contributes to the remarkable decrease in the transcription downstream of the first gene of the frl operon.
INTRODUCTION
Amadori products (fructosamines), the first stable intermediates of the Maillard reaction, result from nonenzymatic glycation of amino acids or proteins with reducing sugars such as glucose. As Amadori products are found in heated food and cause several diseases in connection with diabetes and aging (4), enzymes catalyzing the degradation of Amadori products are of interest to industry and medicine for food processing and diagnostic purposes.
There are two enzymatic mechanisms for the deglycation of Amadori products (reviewed in reference 7): fructosyl-amino acid oxidases (known as Amadoriases) and fructosamine kinases. The former deglycate by means of oxidation, generating the corresponding amino acid, glucosone, and H2O2. Amadoriases have been found in Aspergillus and Penicillium spp. (38, 42) but also in bacteria such as Arthrobacter and Corynebacterium spp. (9, 32). Fructosamine kinases phosphorylate the Amadori products prior to cleavage. Mammalian enzymes add the phosphate at C-3, and the product subsequently undergoes autocatalytic degradation (20). However, Escherichia coli and Bacillus subtilis kinases phosphorylate at C-6; further processing of such intermediates needs a second deglycase enzyme catalyzing the cleavage of the fructosamine 6-phosphates to generate, for example, glucose 6-phosphate and a free amine.
E. coli grows on fructosyl-lysine as the sole carbon and nitrogen source, and expression of frlD (encoding the kinase) and frlB (encoding the deglycating protein) is induced by fructosyl-lysine (39). In B. subtilis, frlD and frlB belong to a gene cluster which additionally comprises three genes coding for putative transporters (frlONM) as well as the convergently oriented putative repressor gene frlR (Fig. 1). Substrate specificities of FrlD and FrlB are quite different than those of the enzymes of E. coli, as B. subtilis acts on α-glycated amino acids rather than on ε-glycated lysine (catalytic efficiencies >30-fold higher), the latter being the preferred substrate of the E. coli enzymes (40). However, other than the biochemical function of the enzymes, little is known about the encoding genes in B. subtilis; neither their physiological significance nor their regulation has been studied in detail.
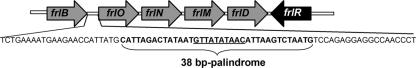
Fig. 1.
Schematic illustration of the frl operon with the frlB and frlO intergenic region. The 38-bp perfect palindromic sequence is shown in bold letters. A putative FrlR binding site (underlined) is located in the center of the intergenic region.
The promoter upstream of the first open reading frame, frlB, was identified as responsive to CodY (26), which is a global transcriptional regulator that controls the expression of more than 100 mostly late-exponential-phase genes in B. subtilis, predominantly as a repressor (reviewed in reference 34). The direct interaction of CodY with the frlB promoter and a strong derepression in a codY deletion strain was revealed (3).
With the recent revision of the B. subtilis 168 genome annotation, a putative regulator gene, frlR, with similarities to the respective gene in E. coli, was identified (2). The predicted FrlR protein was classified as a member of the GntR superfamily, which includes more than 8,500 proteins widely distributed throughout bacteria (reviewed in reference 17). At their conserved N termini, GntRs display helix-turn-helix (HTH) domains; the C terminus routinely facilitates effector binding and/or oligomerization (14). GntRs control a number of fundamental processes, such as catabolism, anabolism, motility, development, antibiotic production, and virulence (5, 8, 12, 15, 18, 19). Based on differences within the C termini and the recognition sequences, seven GntR subfamilies (FadR, HutC, MocR, YtrA, DevA, PlmA, and AraR) have been defined. According to the crystal structure, FrlR belongs to the HutC subfamily (28).
In this study, we investigated the physiological importance of the frl operon in Bacillus subtilis and inspected its regulation by FrlR.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. E. coli XL10-Gold (Stratagene, La Jolla, CA) was used for routine cloning, and E. coli BL21 (37) was used for heterologous protein expression. E. coli strains were grown in Luria-Bertani (LB) broth (33). B. subtilis strains were grown in LB medium or M9 medium supplemented with 1 mg/ml Trp with the appropriate carbon and nitrogen sources. For cultivation in Amadori product, medium M9 was supplemented with 30% (vol/vol) of a 7.5% (wt/vol) Amadori crude preparation (see below). In control medium (M9-Glc-NH4), the equivalent amount of glucose (7.5 g/liter) and, as a nitrogen source, NH4Cl (2.5 g/liter) were used. As a second control medium, the equivalent amount of arginine was used as the sole carbon and nitrogen source (17.7 g/liter). The growth was monitored by measuring the optical density at 600 nm (OD600). When required, the following antibiotics were added with the indicated final concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml in E. coli and 7.5 μg/ml in B. subtilis; erythromycin, 0.5 μg/ml; chloramphenicol, 0.5 μg/ml; tetracycline, 15 μg/ml; and phleomycin, 1 μg/ml.
Table 1.
Strains used in the study
| Strain | Genotypea | Source or reference |
|---|---|---|
| E. coli | ||
| XL10-Gold | endA1 glnV44 recA1 thi-1 gyrA96 relA1 lac Hte Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 Tetr [F′ proAB lacIqZΔM15 Tn10 (Tetr Amy Cmr)] | Stratagene |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | 37 |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| DE01 | trpC2 ΔfrlBONMD ΔfrlR::Ermr | This work |
| DE02 | trpC2 ΔfrlR::Ermr | This work |
| DE11 | trpC2 ΔlacA::BleramyE::[Δ(PfrlB-lacZ) Catr] | This work |
| DE12 | trpC2 ΔfrlR::Ermr ΔlacA::BleramyE::[Δ(PfrlB-lacZ) Catr] | This work |
| DE13 | trpC2 ΔcodY::Kanr ΔlacA::BleramyE::[Δ(PfrlB-lacZ) Catr] | This work |
| DE14 | trpC2 ΔfrlR::Ermr ΔcodY::Kanr ΔlacA::BleramyE::[Δ(PfrlB-lacZ) Catr] | This work |
| DE16 | trpC2 ΔlacA::BleramyE::[Δ(PfrlR-lacZ) Catr] | This work |
| DE20 | trpC2 ΔlacA::BleramyE::[Δ(PfrlB-frlB-IfrlB-frlO-lacZ) Catr] | This work |
| DE21 | trpC2 ΔfrlR::Ermr ΔlacA::BleramyE::[Δ(PfrlR-lacZ) Catr] | This work |
| DE22 | trpC2 ΔfrlR::Ermr ΔlacA::BleramyE::[Δ(PfrlB-frlB-IfrlB-frlO-lacZ) Catr] | This work |
| SK1 | trpC2 ΔlacA::BleramyE::[Δ(PhpaII-frlB-IfrlB-frlO-lacZ) Catr] | This work |
| SK2 | trpC2 ΔlacA::BleramyE::[Δ(PhpaII-lacZ) Catr] | This work |
| SK3 | trpC2 ΔfrlR::Ermr ΔlacA::BleramyE::[Δ(PhpaII-frlB-IfrlB-frlO-lacZ) Catr] | This work |
| SK4 | trpC2 ΔfrlR::Ermr ΔlacA::BleramyE::[Δ(PhpaII-lacZ) Catr] | This work |
| SK7 | trpC2 ΔlacA::BleramyE::[Δ(PfrlB*-lacZ) Catr] | This work |
Amy, amylase expression.
Construction of expression vectors.
Plasmids and oligonucleotides used in this study are listed in Table 2 and Table S1 in the supplemental material, respectively. Standard procedures were used for DNA work (33).
Table 2.
Plasmids used in the study
| Plasmid | Description/genotypea | Source or reference |
|---|---|---|
| pUC18 | Cloning vector (Ampr) | 41 |
| pET26b+ | Vector enabling IPTG-inducible expression of gene of interest with 6× C-terminal His tag (Kanr) | Novagen, Madison, WI |
| pDH32M | Integration vector for transcriptional fusions with lacZ at the amyE locus (Ampr Cmr) | 22 |
| pVD23 | pUC18 derivative containing homologous regions upstream of frlB and downstream of frlR flanking the Ermr gene (Ampr Ermr) | This work |
| pVD24 | pUC18 derivative containing homologous regions upstream and downstream of frlR flanking the Ermr gene (Ampr Ermr) | This work |
| pVD26 | pUC18 derivative containing homologous regions upstream and downstream of codY flanking the Kanr gene (Ampr Kanr) | This work |
| pVD28 | Fusion of pBC16-vector backbone and IPTG-inducible expression cassette of pHT01 (Mobitec) (Tcr) | This work |
| pVD30.1 | pET26b+ derivative containing codY gene (Kanr) | This work |
| pSKL3 | pET26b+ derivative containing frlR gene (Kanr) | This work |
| pVD33.1 | pVD28 derivative containing frlR gene | This work |
| pVD34 | pUC18 derivative containing homologous regions upstream and downstream of lacA flanking the Bler gene (Ampr Bler) | This work |
| pVD36 | pDH32M derivative containing the frlB promoter (Ampr Cmr) | This work |
| pVD37 | pDH32M derivative containing the frlR promoter (Ampr Cmr) | This work |
| pVD39 | pDH32M derivative containing the frlB promoter followed by the frlB gene and IfrlB-frlO (Ampr Cmr) | This work |
| pSKL5 | pDH32M derivative containing the hpaII promoter followed by the frlB gene and IfrlB-frlO (Ampr Cmr) | This work |
| pSKL6 | pDH32M derivative containing the hpaII promoter (Ampr Cmr) | This work |
IPTG, isopropyl-β-d-thiogalactopyranoside.
For amplification of codY and frlR from B. subtilis 168 DNA, the primer pairs codY_NdeI_for/codY_XhoI_rev and frlR_NdeI_for/frlR_XhoI_rev were used and inserted into XhoI/NdeI-digested pET26b+.
The Bacillus expression vector pVD28 was constructed by amplification of the vector backbone of a pBC16 derivative with pVD19_EcoR_for and pVD19_XhoI_rev and amplification of the expression cassette from pHT01 by using the primers Pgrac_EcoRI_for and Pgrac_XhoI_rev. The amplification products were cut with EcoRI and XhoI and subsequently ligated.
Construction of B. subtilis knockout mutants.
For the deletion of genes in B. subtilis, flanking regions of the respective genes were fused to an antibiotic resistance gene by overlap extension PCR (16), and the resulting cassette was cloned into pUC18. The obtained vectors were introduced into B. subtilis 168 by means of natural competence. Double recombination events via the flanks generated mutants selectable by antibiotic resistance; mutant genotypes were confirmed by PCR analysis and sequencing.
For replacement of frlR, the flanks were amplified by using the primers FlA-KpnV/FlA_SOE_Erm and FlB_yurK_EcoR/Erm SOE FlB_yurK and the erythromycin resistance gene with the primers FlA_SOE_Erm/FlB_yurK_SOE_Erm. For disruption of codY, the flanking regions were amplified with primer pairs CodY_FlA-KpnV/Kan_SOE_FlA_codY and CodY_FlB_EcoR/Kan SOE FlB_codY and a kanamycin resistance gene with the primers FlA_codY_SOE_Kan/FlB_codY_SOE_Kan. For deletion of the entire frlBONMD operon along with the regulator gene frlR, homologous regions were amplified by using the primers FlA-KpnV/Erm_SOE_FlA and FlB_EcoR/Erm SOE FlB. The erythromycin resistance gene was amplified using the primers FlA_SOE_Erm and FlB_SOE_Erm. To enable proper β-galactosidase reporter gene assays, the encoding lacA gene was deleted by using primer pairs LacA_FlA-KpnV/ble_SOE_FlA_lacA and FlB LacA_EcoR/ble SOE FlB_lacA for the flanks and FlA_lacA_SOE_ble/FlB_lacA_SOE_ble for the bleomycin resistance gene.
Construction of promoter test strains.
The promoter fragments were amplified while concomitantly generating EcoRI and BamHI restriction sites and cloned into EcoRI/BamHI-digested pDH32M (22). For PfrlB amplification, the primers P-frlB-EcoRI_for/Pfrl_pMUTIN_BamHI_rev were used. PfrlR was amplified with P-frlR-EcoRI_for/P-frlR-BamHI_rev and PhpaII by using PhpaII-BamHI_for/PhpaII_SnaBI_rev. For the amplification of the elongated promoter fragments of PfrlB, the frlB gene, and the frlB-frlO intergenic region (IfrlB-frlO), the primer pair P-frlB-EcoRI_for/P-frlO-BamHI_rev was used. The construction of the elongated PhpaII with the frlB gene and IfrlB-frlO, PCR was carried out using the primer pairs PhpaII-BamHI_for/frlB_SOE_PhpaII and PfrlO SnaBI_rev/PhpaII_SOE_frlB, and a fusion of the amplification products was carried out using overlap extension PCR (16).
After introducing the DNA by means of natural competence, the promoter test mutants were first screened for chloramphenicol resistance and subsequently tested for an amylase-negative phenotype, as the promoter test cassettes were targeted to the amyE locus.
Transformation of B. subtilis by means of natural competence.
B. subtilis cells were transformed by means of natural competence following a modified protocol (1); 500-μl aliquots of competent cells were mixed with various amounts of plasmid DNA (from 1 μg to 10 μg) and incubated for 30 min. For the regeneration of cells, 300 μl LB medium was added and incubated for 30 min. Cells were then plated on LB agar supplemented with the respective antibiotic.
Synthesis of Amadori products.
Amadori products were synthesized by using a modified protocol (10). Glucose and arginine were mixed in a molar ratio of 1:2 in methanol and heated (65.5°C) for 10 to 14 h with stirring and continuous reflux cooling. Subsequently, the methanol was removed by using a rotary evaporator; the obtained product mixture was dissolved in double-distilled water to yield a final concentration of 7.5% (wt/vol).
Determination of glucose, arginine, and Amadori products.
Glucose was determined with the glucose assay reagent of Sigma-Aldrich (St. Louis, MO) as suggested by the manufacturer.
Arginine was measured by ultraperformance liquid chromatography (UPLC) analysis with X-LC (Fasco, Gross-Umstadt, Germany). Arginine was derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate and separated with a Gold Turbo Basic C8 column (Fasco, Gross-Umstadt, Germany).
Relative amounts of Amadori products were measured with the GlyPro reagent of Genzyme Diagnostics (Cambridge, MA). Thus, Amadori products were quantified relative to an internal calibration standard provided with the kit, with a distinct concentration of glycated hemoglobin. Hence, the Amadori amount is specified as a “relative concentration,” since it relates to the glycated hemoglobin control. Twenty-microliter volumes of each sample and of the calibration control (repeated at least twice) were added to 250 μl reagent 1 (R1) and incubated for 5 min at 37°C before measuring the OD550 (A1). Subsequently, 50 μl of reagent 2 (R2) was added, and after an additional 5 min at 37°C, the OD550 (A2) was measured. With the aid of the calibration control the (relative) Amadori product concentration (μmol/liter) was calculated by the equation (ΔAsample/ΔAcalibrator) × 450 μmol/liter, where ΔA = A2 − A1.
RNA isolation and quantitative PCR (qPCR).
B. subtilis 168 was cultivated in LB broth with the addition of 3% (wt/vol) Amadori product, and cells from 7 ml were harvested (4°C) during both exponential growth and stationary growth. Cell samples were added to 3.5 ml of precooled killing buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 20 mM NaN3) and pelleted. After being washed with 5 ml lysis buffer (3 mM EDTA, 200 mM NaCl), the cells were resuspended in 700 μl lysis buffer and transferred into lysing matrix B tubes (Q-Biogene, Montreal, Quebec, Canada) containing 400 μl acidic phenol. Cell disruption was performed with the FastPrepTM FP 120 cell disrupter (BIO 101 Systems, Q-Biogene, Montreal, Quebec, Canada) for 35 s at 6.5 m/s. After centrifugation (12,000 × g for 30 min at 4°C), 350 μl of the upper phase was transferred into a fresh tube, and the RNA was purified with the aid of a KingFisher instrument (Thermo Electron Corporation; Cambridge, MA) and MagNa Pure LC RNA isolation kits (Roche Diagnostics AG, Rotkreuz, Switzerland) by applying the protocol suggested by the manufacturer. A DNA digestion step with RQ1 RNase-free DNase (Promega GmbH, Mannheim, Germany) was subsequently carried out, and the RNA concentration was determined with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). The quality and purity of RNA were determined by using an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, CA) and an RNA 6000 NanoLab chip kit (Agilent Technologies Inc., Santa Clara, CA); total RNA was transcribed into cDNA by applying a high-capacity cDNA reverse transcription kit (Applied Biosystems, CA) with the protocol recommended by the manufacturer.
For the relative quantitative determination of frlB and frlD transcripts, qPCRs were performed in an MXPro 3500 system (Stratagene, La Jolla, CA) with the Brilliant SYBR green qPCR master mix kit (Stratagene, La Jolla, CA). A total of 0.032 ng cDNA was used per well, and the analysis was carried out in triplicate; the primers used for measurement of frlB and frlD transcripts were qPCR-yurP-2-for/qPCR-yurP-2-rev and qPCR-yurL-5-for/qPCR-yurL-5-rev, respectively. Primer design was done with the eprimer3 software (EMBOSS [the European Molecular Biology Open Software Suite] package). For the housekeeping gene atpA, which served as a reference, the primers qPCR-atpA-3-for/qPCR-atpA-3-rev were used. The software for the MXPro 3500 system (Stratagene, La Jolla, CA) was applied for the determination of threshold cycle (CT) values, and ΔCT values were calculated in relation to the value for atpA.
Expression and purification of FrlR and CodY proteins.
E. coli BL21(DE3) expression strains containing pVD30.1 or pSKL3 in the pET26b+ (Novagen, Madison, WI) plasmid including a C-terminally 6-histidine-tagged codY gene (pVD30.1) or frlR gene (pSKL3) were grown in autoinduction medium [50 mM NaH2PO4 · 1H2O, 50 mM KH2PO4, 25 mM (NH4)2SO4, 1 mM MgSO4 · 7H2O, 0.4% (wt/vol) glycerin, 3 mM glucose, 6 mM lactose, 10 g/liter NZ-Amine-AS, 5 g/liter Bacto yeast extract, 20 μM CaCl2 · 2H2O, 10 μM MnCl2 · 4H2O, 10 μM ZnCl2, 2 μM CoCl2, 2 μM CuCl2, 2 μM NiCl2, 2 μM Na2MoO4 · 2H2O, 2 μM Na2SeO3, 2 μM H3BO3, 50 μM FeCl3 · 6H2O] supplemented with kanamycin (50 μg/ml). Cultivation was carried out at 30°C and 200 rpm (incubator ISF-1-W; Kühner, Birsfelden, Switzerland). After 24 h, cells were pelleted and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole), the cells were disrupted by sonication (amplitude of 40% for 2.5 min, with a time interval of 5 s and an S&M 0102 tube in a Sonics & Materials Inc. Vibra Cell [Meyrin/Satigny, Switzerland]), and the debris was removed by centrifugation. The 6×His-tagged CodY was purified using Ni-nitrilotriacetic acid (NTA) agarose obtained from Invitrogen (Darmstadt, Germany). The protein concentration in the eluates was measured using the Pierce bicinchoninic acid (BCA) protein assay kit (Pierce Biotechnology, Rockford, IL) by using bovine serum albumin (BSA) as a relative standard as recommended by the manufacturer.
Electrophoretic mobility shift assay (EMSA).
Five femtomoles of 5′-digoxigenin (DIG)-labeled promoter fragments were incubated with increasing concentrations of C-terminally 6×His-tagged FrlR (FrlRcHis) in a 10-μl reaction mixture at 22°C for 20 min. The reaction mixture contained 20 mM Tris-HCl (pH 8.0), 50 mM sodium glutamate, 10 mM MgCl2, 5 mM EDTA, 5% (vol/vol) glycerol, and 250 ng calf thymus DNA. Directly after incubation, 2 μl of a 5-times-concentrated sample buffer (18 mM Tris, 18 mM boric acid, 0.4 mM EDTA, 9% Ficoll type 400, 0.02% bromphenol blue) was added and the samples were loaded onto a 6% DNA retardation gel (Invitrogen, Darmstadt, Germany) running at 50 V and 4°C for 90 min. The DNA was electrotransferred (30 V for 90 min) to a positively charged nylon membrane (Porablot NY Plus nylon membrane, 0.45-μm pore size; Macherey-Nagel, Düren, Germany) using 0.5× Tris-borate-EDTA (TBE) as the transfer buffer. The immunological detection of the 5′-DIG-labeled promoter fragments was performed with an antidigoxigenin antibody (Roche, Mannheim, Germany) and nitroblue tetrazolium (NBT)-BCIP (5-bromo-4-chloro-3-indolylphosphate) solution as recommended by the manufacturer.
Promoter test analysis by o-nitrophenyl-β-d-galactopyranoside (ONPG) assay.
Promoter test mutants were cultivated in 100-ml Erlenmeyer flasks (300 rpm in incubator ISF-1-W; Kühner, Birsfelden, Switzerland) (37°C) in a total volume of 10 ml medium. Reporter gene analyses were carried out as described previously (23) with the following equation: Miller units (MU) = (OD420 × 1,000)/(OD600 × V × t) (23).
RESULTS
The frl gene cluster is essential for Amadori product degradation.
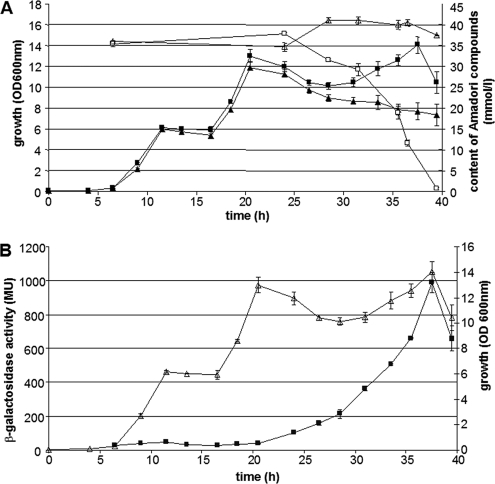
As we cannot exclude the possibility that B. subtilis possesses enzymes other than those encoded by the frl operon capable of degrading fructosamines, the physiological significance was investigated by generating a ΔfrlBONMDR mutant. The growth and Amadori product consumption of the deletion mutant were compared with those of the wild-type strain (Fig. 2A). The growth of the wild type in a medium with crude preparations of the Amadori product fructosyl-arginine (containing residua of glucose and arginine) displays three phases. Glucose, arginine, and fructosyl-arginine were monitored by using enzymatic assay kits for glucose and Amadori products and UPLC analysis for arginine. The initial growth phase resulted from glucose consumption followed by arginine consumption, and finally the Amadori product was consumed. The lack of the last growth phase in the deletion mutant is consistent with the remaining constant concentration of Amadori products and demonstrates the inability of the ΔfrlBONMDR strain to use fructosylamine. Thus, the genes of the frlBONMDR cluster appear to be instrumental in and necessary for Amadori product degradation. As the ΔfrlR mutant degraded the fructosamines (see Fig. S1 of the supplemental material), frlR is not necessary for growth on Amadori products, whereas some or all of the genes of the operon appear to be necessary.
Fig. 2.
Growth of B. subtilis in M9 Amadori product medium. (A) Comparison of the growth profiles of wild type (squares) and DE01 (ΔfrlBONMD ΔfrlR; triangles). The OD600s were determined as a parameter for growth (filled squares and triangles), and the relative concentrations of Amadori products in culture supernatant (open squares and triangles) were quantified by use of GlyPro reagents (Genzyme) relative to a glycated hemoglobin calibrator control. (B) The expression of the lacZ reporter gene driven by PfrlB of wild-type strain DE11 (filled squares) during growth in M9-Amadori product medium (open triangles). Standard deviations are shown as error bars.
FrlR is a negative regulator of the frlBONMD operon.
Results of reporter gene assays performed with a PfrlB-lacZ fusion integrated into the amyE locus (strain DE11; Fig. 2B) demonstrate a strong increase in expression from PfrlB during the growth on Amadori products. As outlined in the introductory remarks, the expression of the frl genes driven by PfrlB is under the control of the global regulator CodY, which represses transcription from PfrlB during growth on glucose (3). Based on the results outlined in Fig. 2, another transcriptional regulator besides CodY may participate; thus, we checked the possible but hitherto unknown involvement of the putative regulator FrlR, which is encoded adjacent to the structural frlBONMD genes (see the schematic representation in Fig. 1).
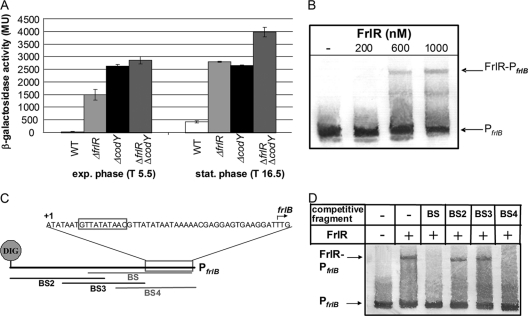
DE12 and DE13 (FrlR and CodY deletion strains, respectively) were constructed by replacing frlR and codY with erythromycin and kanamycin resistance genes, respectively (the double mutant was DE14) and the expression of the PfrlB-lacZ fusion was compared to the wild-type expression. Results obtained with the ΔcodY strain, DE13, agree with those of Belitsky and Sonenshein (3), as the mutant displayed constant high-level β-galactosidase activities (Fig. 3A). Consistently, in our Amadori product medium, DE13 displayed constant high-level reporter activities already in the first growth phase (on residual glucose); however, the activity levels doubled in second growth phase (data not shown). The reporter gene assay of the frlR deletion strain (DE12) disclosed a similar strong repression of PfrlB by FrlR (Fig. 3A). The frlR mutation enhanced the β-galactosidase activity approximately 50-fold during the exponential growth phase and also 7-fold in stationary phase when grown in glucose medium. Such repression is at least partially independent of CodY, as the double mutant DE14 (ΔfrlR ΔcodY) was further derepressed in the stationary growth phase compared to the single mutants (about 100-fold; Fig. 3A).
Fig. 3.
(A) Measurement of the frlB promoter activity in the strains DE11 (wild type [WT]), DE12 (ΔfrlR), DE13 (ΔcodY), and DE14 (ΔfrlR ΔcodY), quantified as lacZ reporter gene activities in Miller units. Cells were grown in M9-Glc-NH4 medium. Samples were taken at 5.5 h after inoculation (T5.5; exponential [exp.] growth phase) and T16.5 (stationary [stat.] phase). Levels for DE11 (WT) kept constant until the culture reached the stationary phase; maximum activity in the stationary phase is given in the figure and kept at this level until lysis occurred. Standard deviations are shown as error bars. (B) Analysis of binding of FrlR to PfrlB by EMSA. A volume of 0.5 nM 5′-labeled PfrlB was incubated without (−) or with 200 to 1,000 nM FrlRcHis. (C) Schematic illustration of applied unlabeled fragments of the frlB promoter for competitive EMSA experiments. The consensus GntR binding site is boxed, and the transcriptional start site (+1) is indicated as such (3). The CodY binding site (3) is located in fragment BS3. (D) Electrophoretic mobility shift assay with 5′-DIG-labeled PfrlB and different single competitor fragments in 20-fold excess. The amount of unlabeled competitors added to each reaction mixture was 10 nM (20× relative to the labeled PfrlB). The amount of 5′-DIG-labeled probe PfrlB was 0.5 nM. The concentration of FrlRcHis was 800 nM.
The direct binding of FrlR to PfrlB was investigated by performing electrophoretic mobility shift assays (EMSAs). The entire intergenic region (199 bp) upstream of frlB was incubated with various concentrations of the FrlR protein, which was heterologously expressed in E. coli with a His tag for ease of purification and of immunological detection. In PAGE, a distinct shift of the promoter fragment upon the addition of 600 nM FrlR became visible (Fig. 3B).
In order to localize the binding site of FrlR within the frlB promoter sequence, competitive mobility shift assays were performed. Four partially overlapping fragments of the frlB promoter were generated (Fig. 3C; see Fig. S4 in the supplemental material) and individually incubated in 20-fold excess with the complete labeled promoter fragment and the FrlR protein. Binding of FrlR to the competing fragments reduces the binding of FrlR to the labeled promoter fragment and, thus, decreases the amount of the shifted EMSA fragment. As shown in Fig. 3D, the decrease of the shifted fragment occurs with competitor DNA fragments BS and BS4 (Fig. S4), thus localizing the FrlR binding site to the overlapping regions of the two fragments. Within the identified region, GntR regulator type consensus sequence 5′-(N)yGT(N)xAC(N)y-3′ (where y is the number and N is the nature of the nucleotides being palindromic around the conserved GT-AC which flank several A/T bases [x]) is recognizable. In the frlB promoter region, a 26-bp palindrome with a central GTtaTAtaAC sequence (with capital letters representing conserved nucleotides) suggests that FrlR belongs to the HutC subfamily of the GntRs, as these are characterized by a central TA between the conserved GT and AC bases (30).
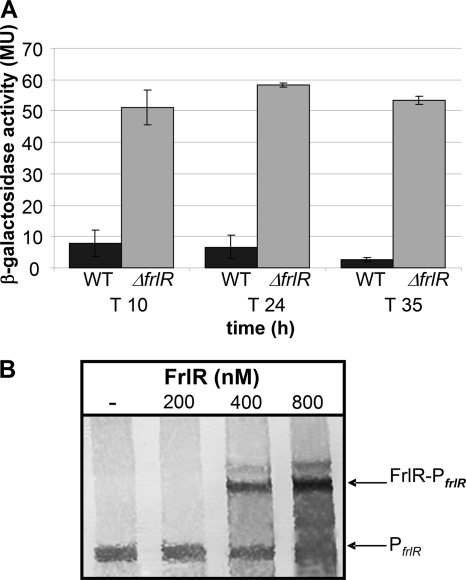
Expression of frlR is subject to autoregulation.
To test possible autoregulation, a strain carrying PfrlR upstream of the lacZ gene was constructed. The expression of the reporter gene in the wild-type background (DE16) was rather low (∼3 to 10 MU, depending on the growth phase and the medium). To further examine potential autoregulation, which is not uncommon for GntR regulators (31), the strain carrying the frlR deletion was examined. The PfrlR-lacZ expression in the ΔfrlR strain was increased about 5- to 10-fold compared to that of the wild type (Fig. 4A). Subsequently, the direct binding of FrlR to its own promoter was tested by mobility shift assays. The shift of the promoter fragment upon the addition of 400 nM FrlR repressor (Fig. 4B) revealed the binding of FrlR, which agrees with negative transcriptional autoregulation.
Fig. 4.
Investigation of autoregulation of frlR expression. (A) The frlR promoter activities in strains DE16 (wild type) and DE21 (ΔfrlR) were determined as lacZ reporter gene activities in Miller units. Cells were grown in M9-Amadori product medium, and samples were taken at three different time points. Standard deviations are shown as error bars. (B) Examination of FrlR binding to PfrlR by EMSA was carried out with 0.5 nM 5′-DIG-labeled PfrlR and various concentrations of FrlRcHis (200 to 800 nM).
The first intergenic region of the operon mediates the decrease in expression of the downstream genes.
In qPCR analyses, atpA, a constantly expressed housekeeping gene, was used as a reference. The PCR analyses validated the reporter gene assays described above; however, though the two genes belong to the same transcription unit, expression of frlB was ca. 4-fold higher than that of frlD (data not shown). This agrees with recent tiling arrays in which expression was found to fade downstream of frlB (27). Hence, our attention was drawn to the first intergenic sequence (IfrlB-frlO) of the operon. We identified a perfect 38-bp-spanning palindrome (Fig. 1) which may form a hairpin with a stem of at least 17 bp, allowing for a 4-bp loop, possibly acting as pausing site for the RNA polymerase. Furthermore, the sequence in the center of the palindrome matches the FrlR binding site GT..AT..AC (Fig. 1), suggesting that FrlR has in addition to the frlB and frlR promoters a third binding site within the cluster. Indeed, EMSA experiments confirmed binding of FrlR to the intergenic region, as a shift was seen at the addition of 500 nM FrlR to the labeled IfrlB-frlO (Fig. 5A).
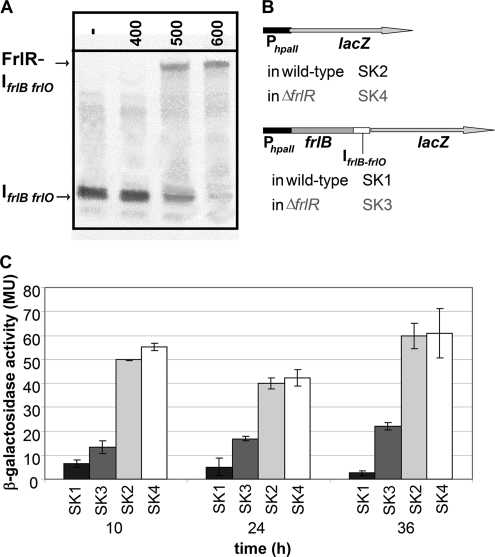
Fig. 5.
The frlB-frlO intergenic region (IfrlB-frlO) decreases the expression downstream of frlB. (A) Analysis of the binding of FrlR to IfrlB-frlO by EMSA. A 0.5 nM concentration of 5′-DIG-labeled IfrlB-frlO was incubated without (−) or with 400 to 600 nM FrlRcHis. (B) Schematic illustration of the reporter gene constructs. The FrlR-independent hpaII promoter was cloned in front of the lacZ reporter gene in wild-type (SK2) and ΔfrlR (SK4) constructs. Further constructs with the frlB gene and IfrlB-frlO downstream of PhpaII followed by lacZ (SK1 [wild type] and SK3 [ΔfrlR]) were generated. (C) Measurement of promoter activity by β-galactosidase assay. Strains were cultivated in M9-Amadori medium, and at the indicated time points, the β-galactosidase activity was measured. Standard deviations are shown as error bars.
Promoter test studies were carried out to examine a possible expression reduction by the palindrome. The frlB gene with the intergenic region IfrlB-frlO was cloned between PfrlB and lacZ to compare its expression to the expression of PfrlB alone. Such a reporter gene construct resembles the native situation, as the start codon of lacZ replaces the frlO start codon. In the wild type, the decreasing effect of the elongated construct with the intergenic region downstream of PfrlB was about 2- to 5-fold in comparison to the PfrlB alone (see Fig. S2 in the supplemental material). Expression from the PfrlB promoter in the ΔfrlR background was much stronger than its expression in the wild type; a direct comparison with the wild type is not possible, as FrlR binds to both PfrlB and IfrlB-frlO.
This binding is why an FrlR-independent promoter was subsequently used to exclude overlapping effects by FrlR binding to the promoter and the intergenic region. The promoter PhpaII of the Staphylococcus aureus pUB110 plasmid was cloned upstream of either the lacZ reporter gene only or the frlB gene, with IfrlB-frlO also placed in between PhpaII and lacZ. A schematic representation of the constructs is depicted in Fig. 5B. Reporter gene expression of these constructs in the wild type and the ΔfrlR background was subsequently inspected (Fig. 5C). Though we cannot exclude the possibility that different junctions between the promoter and lacZ may influence expression of the reporter gene, it is unquestionable that FrlR reduces this expression. Furthermore, the results obtained with constructs resembling the native genetic organization are supportive of our conclusion, that the decreasing effect of the intergenic region is apparent and clearly not dependent on the upstream promoter, as the effects on expression with PhpaII and PfrlB are similar. However, a considerable difference became obvious by using the PhpaII promoter when wild-type and ΔfrlR strains were compared (Fig. 5C), as the decreasing effect is stronger in the wild type, thereby suggesting that FrlR is involved in enforcing the reductive effect on expression.
DISCUSSION
Since the B. subtilis deletion mutant lacking the frlBONMDR cluster failed to consume and grow on fructosamines, the genes of the operon are not only mandatory but evidently also sufficient for the consumption of Amadori products. Moreover, fructosamines can apparently serve as the sole carbon and nitrogen sources in B. subtilis as for E. coli (40).
Our findings that the expression level increased at the end of the exponential growth phase/beginning of the stationary phase even in media with glucose as the sole carbon source agree with previous studies in which the global transcriptional regulator CodY was shown to downregulate the expression of the frl operon during exponential growth (3). However, compared to the corresponding single mutants, the ΔfrlR ΔcodY double mutant displayed a clear increase in expression driven by PfrlB even without Amadori products being present in the medium, which suggested that FrlR acts as an additional repressor besides CodY, which has a known role. Moreover, the derepression in media supplemented with Amadori products was greater than that in glucose medium (16-fold in glucose medium versus 33-fold in fructosamine medium). Thus, in the wild-type strain, expression is almost totally repressed during exponential growth on glucose and partially derepressed after the exponential growth; however, full derepression requires the presence of Amadori products. During growth on arginine, repression of the wild-type strain is constant and on a maximum level even at the stationary phase. Under the presumption that fructosamines serve as effector molecules, the strong derepression during growth on fructosamines could be understood. However, Amadori products may not be the only effectors, since during growth on glucose, half-maximal derepression (16-fold in comparison with 33-fold) was demonstrated. Interestingly enough, within the crystal structures of TreR, YvoA, and FrlR, there is a sulfate ion at the same location, which is characterized by conserved residues for sulfate binding (28). For TreR, it has been proposed that sulfate mimics the phosphoryl group of the effector trehalose 6-phosphate (29), and YvoA is inactivated by its cognate effector N-acetylglucosamine-6-phosphate (28).
This agrees with the finding that intracellular glycation frequently occurs in E. coli, baker's yeast, and mammals (6, 13, 24, 25, 35, 36). Moreover, the glycating potential of glucose 6-phosphate is 3- to 20-fold higher (depending on the amine) than that of glucose (11, 13, 35, 36). Hence, fructosamine 6-phosphate, rather than fructosamine, is likely to act as the actual (or as an additional) effector.
An alignment of the FrlR binding sites (Fig. 6) revealed the conserved operator motif (GT..TA..AC) of the HutC subfamily of GntR regulators (30) flanked by conserved palindromic sequences of different lengths, and indeed, preliminary mutation studies (with replacements of the conserved G and C with A and T within the frlB promoter) revealed a partial loss of FrlR binding in EMSA experiments and reporter gene assays (see Fig. S3 in the supplemental material). The deletion of frlR did not lead to a noticeable phenotype, but overexpression eventually was lethal (data not shown), possibly indicating that FrlR is involved in the regulation of other loci, though, due to the resulting large amount of the protein during overexpression, the repressor may also switch off promoters with binding sites displaying a lower affinity. As for E. coli, the glycopeptidase Gcp may be a putative target for FrlR regulation. The enzyme may protect the bacteria from damage caused by Amadori-modified proteins, and indeed, highly conserved Gcp proteins are seen in almost all sequenced organisms (21).
Fig. 6.
Alignment of the identified FrlR binding sites in the frlB promoter, the frlR promoter, and the frlB-frlO intergenic region. Homologous bases are indicated as black letters on a gray background. The consensus motif for GntR regulators of the HutC family is framed in black.
As there was an additive effect of the palindrome and FrlR binding in our reporter studies, we assume that the palindrome may form a hairpin, causing transcription termination; additionally, binding of FrlR may build a roadblock for the RNA polymerase, thereby weakening expression of the downstream genes (27).
Such downregulation of the transporter genes frlONM and of the kinase gene frlD makes sense, taking into account, on one hand, kinetic parameters of both the kinase and the deglycase, with the former displaying considerably higher kcat and kcat/Km values for all tested substrates than FrlB (40). On the other hand, glucose 6-phosphate actually produces significant intracellular glycation, and solely FrlB is needed for their degradation. The transporter encoded by frlONM is probably necessary for the import of fructosamines; thus, expression of the whole operon is required for growth on Amadori products. The frl genes in B. subtilis apparently have two physiological functions: facilitating growth on extracellular Amadori products and protecting cells from damage by removing intracellularly formed fructosamine phosphates.
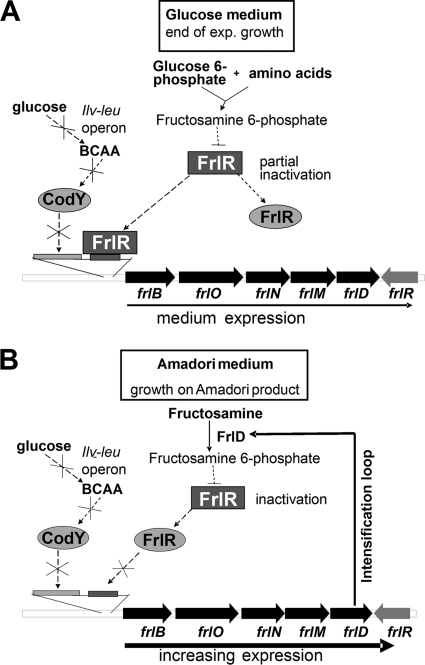
As outlined in Fig. 7, intracellular glucose 6-phosphate may result in glycation of proteins and amino acids (Fig. 7A). Fructosamine 6-phosphates may inactivate FrlR, eventually leading to increased expression of the frl operon. CodY is inactive at the end of the exponential growth phase when glucose is exhausted. During growth on glucose, the CcpA-mediated activation of the ilv-leu operon causes the synthesis of the branched-chain amino acids isoleucine, leucine, and valine acting as activators for CodY.
Fig. 7.
Postulation of the regulation of the frl genes in B. subtilis. (A) During growth on glucose, intracellular glucose 6-phosphate leads to the glycation of proteins and amino acids; such fructosamine 6-phosphates partially inactivate FrlR, thereby promoting expression of the frl operon. CodY is inactive at the end of the exponential growth phase, due to glucose exhaustion. During growth on glucose, the CcpA-mediated activation of the ilv-leu operon causes the synthesis of the branched-chain amino acids (BCAA) isoleucine, leucine, and valine acting as CodY activators. (B) At the beginning of growth on Amadori products, import and phosphorylation of the fructosamines (by FrlD) represent the limiting step, since the frl operon is initially expressed at a low level. The more Amadori products are imported and phosphorylated (with the help of increasing expression of frlONMD), the more FrlR is inactivated, resulting in the further increase of expression, initiating an intensification loop and allowing for positive feedback regulation.
On Amadori products, the import and phosphorylation of the fructosamines (by FrlONM and FrlD, respectively) may be the rate-limiting step, as the frl operon is initially expressed at a low level. The more Amadori products are being imported and phosphorylated (with the help of increasing expression of frlONMD), the more FrlR is inactivated eventually leading to increased expression and, thus, to an intensification loop, i.e., positive feedback regulation (Fig. 7B).
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Anagnostopoulos C., Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbe V., et al. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775 doi:10.1099/mic.0.027839-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belitsky B. R., Sonenshein A. L. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224–1236 doi:10.1128/JB.01780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brownlee M. 1995. Advanced protein glycosylation in diabetes and aging. Annu. Rev. Med. 46:223–234 doi:10.1146/annurev.med.46.1.223 [DOI] [PubMed] [Google Scholar]
- 5. Casali N., White A. M., Riley L. W. 2006. Regulation of the Mycobacterium tuberculosis mce1 operon. J. Bacteriol. 188:441–449 doi:10.1128/JB.188.2.441-449.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colas B., Boulanger Y. 1983. Glycosylation of yeast aspartyl-tRNA synthetase. Affinity labelling by glucose and glucose 6-phosphate. FEBS Lett. 163:175–180 [DOI] [PubMed] [Google Scholar]
- 7. Deppe V. M., Bongaerts J., O'Connell T., Maurer K. H., Meinhardt F. 2011. Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions. Appl. Microbiol. Biotechnol. doi:10.1007/s00253-010-3083-4 [DOI] [PubMed] [Google Scholar]
- 8. DiRusso C. C., Nystrom T. 1998. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 27:1–8 [DOI] [PubMed] [Google Scholar]
- 9. Ferri S., Sakaguchi A., Goto H., Tsugawa W., Sode K. 2005. Isolation and characterization of a fructosyl-amine oxidase from an Arthrobacter sp. Biotechnol. Lett. 27:27–32 doi:10.1007/s10529-004-6312-z [DOI] [PubMed] [Google Scholar]
- 10. Finot P. A., Mauron J. 1969. Le blocage de la lysine par la réaction de Maillard. 1. Synthèse de N-(desoxy-1-D-fructosyl-1) et N-(desoxy-1-D-lactulosyl-1)-L-lysine. Helv. Chim. Acta 52:1488–1495 [DOI] [PubMed] [Google Scholar]
- 11. Fortpied J., Gemayel R., Stroobant V., van Schaftingen E. 2005. Plant ribulosamine/erythrulosamine 3-kinase, a putative protein-repair enzyme. Biochem. J. 388:795–802 doi:10.1042/BJ20041976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haine V., et al. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578–5586 doi:10.1128/IAI.73.9.5578-5586.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haney D. N., Bunn H. F. 1976. Glycosylation of hemoglobin in vitro: affinity labeling of hemoglobin by glucose-6-phosphate. Proc. Natl. Acad. Sci. U. S. A. 73:3534–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haydon D. J., Guest J. R. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291–295 [DOI] [PubMed] [Google Scholar]
- 15. Hillerich B., Westpheling J. 2006. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 188:7477–7487 doi:10.1128/JB.00898-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 17. Hoskisson P. A., Rigali S. 2009. Chapter 1: variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv. Appl. Microbiol. 69:1–22 doi:10.1016/S0065-2164(09)69001-8 [DOI] [PubMed] [Google Scholar]
- 18. Hoskisson P. A., Rigali S., Fowler K., Findlay K. C., Buttner M. J. 2006. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J. Bacteriol. 188:5014–5023 doi:10.1128/JB.00307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaques S., McCarter L. L. 2006. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 188:2625–2635 doi:10.1128/JB.188.7.2625-2635.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kappler F., Schwartz M. L., Su B., Tobia A. M., Brown T. 2001. DYN 12, a small molecule inhibitor of the enzyme amadoriase, lowers plasma 3-deoxyglucosone levels in diabetic rats. Diabetes Technol. Ther. 3:609–616 [DOI] [PubMed] [Google Scholar]
- 21. Katz C., Cohen-Or I., Gophna U., Ron E. Z. 2010. The ubiquitous conserved glycopeptidase Gcp prevents accumulation of toxic glycated proteins. MBio 1:195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kraus A., Hueck C., Gartner D., Hillen W. 1994. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 176:1738–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller J. H. 1972. Experiments in molecular genetics, p. 352–355 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Mironova R., Niwa T., Handzhiyski Y., Sredovska A., Ivanov I. 2005. Evidence for non-enzymatic glycosylation of Escherichia coli chromosomal DNA. Mol. Microbiol. 55:1801–1811 doi:10.1111/j.1365-2958.2005.04504.x [DOI] [PubMed] [Google Scholar]
- 25. Mironova R., Niwa T., Hayashi H., Dimitrova R., Ivanov I. 2001. Evidence for non-enzymatic glycosylation in Escherichia coli. Mol. Microbiol. 39:1061–1068 [DOI] [PubMed] [Google Scholar]
- 26. Molle V., et al. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasmussen S., Nielsen H. B., Jarmer H. 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73:1043–1057 doi:10.1111/j.1365-2958.2009.06830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Resch M., Schiltz E., Titgemeyer F., Muller Y. A. 2010. Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA. Nucleic Acids Res. 38:2485–2497 doi:10.1093/nar/gkp1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rezácová P., et al. 2007. The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacillus subtilis 168 reveals a unique quarternary assembly. Proteins 69:679–682 doi:10.1002/prot.21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rigali S., Derouaux A., Giannotta F., Dusart J. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507–12515 doi:10.1074/jbc.M110968200 [DOI] [PubMed] [Google Scholar]
- 31. Ryding N. J., et al. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343–357 [DOI] [PubMed] [Google Scholar]
- 32. Sakaue R., Hiruma M., Kajiyama N., Koyama Y. 2002. Cloning and expression of fructosyl-amino acid oxidase gene from Corynebacterium sp. 2-4-1 in Escherichia coli. Biosci. Biotechnol. Biochem. 66:1256–1261 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Sonenshein A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 doi:10.1016/j.mib.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 35. Stevens V. J., Rouzer C. A., Monnier V. M., Cerami A. 1978. Diabetic cataract formation: potential role of glycosylation of lens crystallins. Proc. Natl. Acad. Sci. U. S. A. 75:2918–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stevens V. J., Vlassara H., Abati A., Cerami A. 1977. Nonenzymatic glycosylation of hemoglobin. J. Biol. Chem. 252:2998–3002 [PubMed] [Google Scholar]
- 37. Studier F. W., Moffatt B. A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 38. Takahashi M., Pischetsrieder M., Monnier V. M. 1997. Isolation, purification, and characterization of amadoriase isoenzymes (fructosyl amine-oxygen oxidoreductase EC 1.5.3) from Aspergillus sp. J. Biol. Chem. 272:3437–3443 [DOI] [PubMed] [Google Scholar]
- 39. Wiame E., Delpierre G., Collard F., Van Schaftingen E. 2002. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J. Biol. Chem. 277:42523–42529 doi:10.1074/jbc.M200863200 [DOI] [PubMed] [Google Scholar]
- 40. Wiame E., Duquenne A., Delpierre G., Van Schaftingen E. 2004. Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis. FEBS Lett. 577:469–472 doi:10.1016/j.febslet.2004.10.049 [DOI] [PubMed] [Google Scholar]
- 41. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 42. Yoshida N., Sakai Y., Serata M., Tani Y., Kato N. 1995. Distribution and properties of fructosyl amino acid oxidase in fungi. Appl. Environ. Microbiol. 61:4487–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.