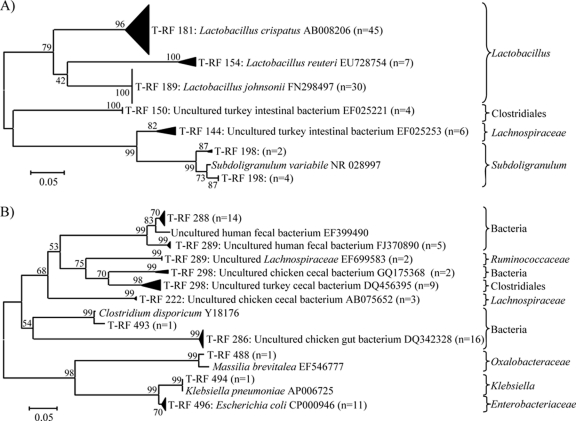
Fig. 2.
Unrooted neighbor-joining phylogenetic tree of 16S rRNA gene sequences (T-RFs) obtained by the targeted cloning and sequencing of OTUs identified as being significantly affected by dietary treatment. (A) Phylogenetic tree from OTUs of less than 200 bp. (B) Phylogenetic tree from OTUs of greater than 200 bp. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The line bars indicate evolutionary distances and are in units of the number of base substitutions per site. The values along the branches indicate percent confidence. The numbers in parentheses indicate numbers of sequences analyzed from multiple samples. The GenBank accession numbers with the closest identity to the T-RFs are indicated. The classifications of T-RF sequences according to the RDP classifier are indicated to the right of the tree.