Figure 4.
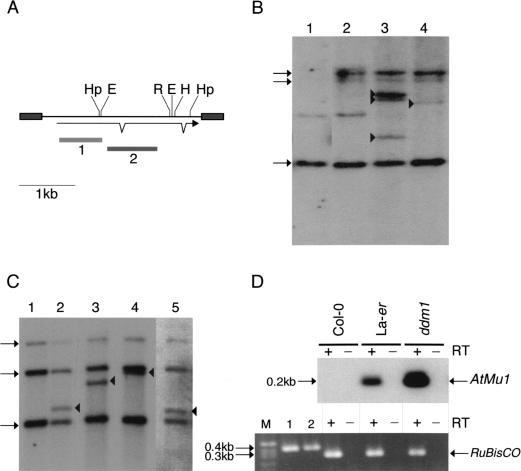
Mutator transposons are activated in ddm1 mutants.(A) AtMu1 has 295 bp TIRs (grey boxes) and encodes three exons; probes 1 and 2 are indicated below. Restriction enzyme sites for HpaII (Hp), HindIII (H), EcoRI (R), and EcoRII (E) are shown. (B) DNA gel blot analysis of pooled wild-type and ddm1-mutant seedlings. DNA was digested with EcoRI and hybridized with probe 2. New bands (arrowheads) represent transposition of AtMu1. The preexisting elements T11I11.3, F21A20_a, and T3F12.12 are indicated by arrows on the left. The faint band in lanes 1 and 2 is partially methylated T3F12.12. Lane 1, Landsberg erecta (Ler); lane 2, Columbia wild-type (Col-0); lane 3, Columbia ddm1; lane 4, Landsberg erecta ddm1. (C) DNA gel blot analysis of individual Columbia ddm1 plants using HindIII and probe 1. New bands (arrowheads) are transposed AtMu1. (D) RT–PCR analysis (+) of transcripts using AtMu1 primers (top panel) and control RuBisCO primers (bottom panel). Alternate lanes (−) are mock reactions in the absence of reverse transcriptase. RuBisCO lanes 1 and 2 were amplified using Col-0 and Ler genomic DNA.