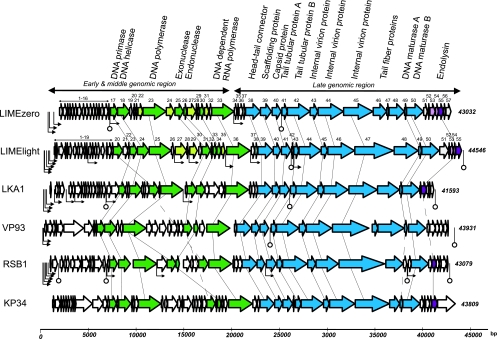
Fig. 2.
Comparison of the genomes of Pantoea phages LIMEzero and LIMElight with those of Pseudomonas phage LKA1, Vibrio phage VP93, Ralstonia phage RSB1, and Klebsiella phage KP34. The predicted open reading frames are indicated by arrows. ORFs of unknown function in the Pantoea phages are in light gray, predicted DNA metabolism genes in green, structural genes in blue, and lysis genes in purple. (ORFs in lighter shades have no equivalents in the other phages.) Functionally equivalent genes are connected by thin lines, and putative functions are designated. Host promoters are indicated by bent arrows with triangular points and phage-specific promoters by smaller bent arrows; factor-independent terminators are shown with stem-loop structures.