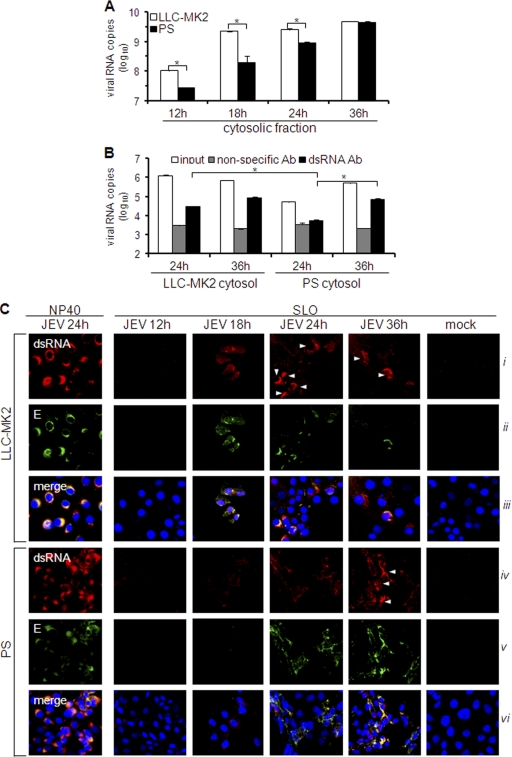
Fig. 8.
Quantitation of cytosolic viral RNA and double-stranded RNA. (A and B) LLC-MK2 or PS cells were infected with JEV (MOI, 1), and cytosolic fractions were obtained at various times postinfection. (A) RNA was isolated from cytosolic extracts using the spin column method and subjected to viral RNA quantification by real-time RT-PCR. The results are expressed as log10 copies of viral RNA per 1 × 106 cells. The error bars indicate standard deviations of the means. The asterisks indicate the statistical significance (P < 0.001) between the comparison groups. (B) Phenol-extracted RNA from the cytosolic extract was incubated with nonspecific or dsRNA-specific antibody on immunosorbent plates or left untreated (input). Antibody-bound RNA was harvested by proteinase K/SDS treatment, reextracted, and quantified by real-time RT-PCR using JEV-specific primers. The results are expressed as log10 copies of viral RNA per 100 ng total RNA. The error bars indicate standard deviations of the means. The asterisks indicate the statistical significance (P < 0.001) between the comparison groups. (C) Immunodetection of dsRNA and E protein in NP-40- or SLO-permeabilized cells at various times postinfection by JEV (MOI, 1). NP-40 was used to permeabilize all membranes or SLO to permeabilize only the plasma membrane. Nuclear staining was achieved using DAPI. The white arrowheads indicate cells with intense dsRNA staining.