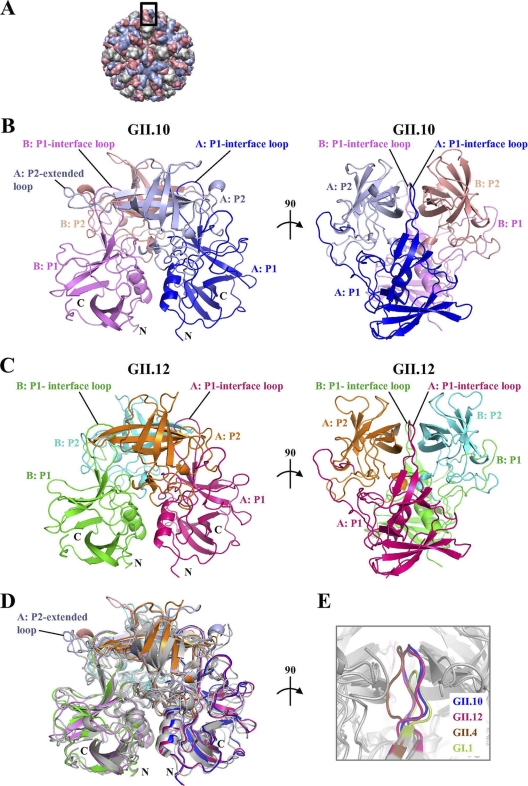
Fig. 1.
Structures of the norovirus GII.10 and GII.12 P domains. The GII.10 and GII.12 P1 subdomains are very similar, with greater differences observed in the P2 subdomains. (A) The GII.10 VLP was modeled from the shell domain of the norovirus (NV) VLP (PDB ID, 1IHM) and the unbound GII.10 P domain (PDB ID, 3ONU). The GII.10 VLP (T = 3) was modeled with different monomer interactions, A/B and C/C, where each A, B, and C monomer was colored light blue, salmon, and orange, respectively. The boxed region showed the location of the P domain capsid dimer. (B) The X-ray crystal structure of the unbound GII.10 P domain dimer was determined to have 1.4-Å resolution and colored according to monomers (chains A and B) and P1 and P2 subdomains, i.e., chain A P1 (blue), chain A P2 (light blue), chain B P1 (violet), and chain B P2 (salmon). The chain A P2-extended loop protruded out from the side of the P domain (the chain B extended loop was not fitted into the structure). The P1-interface loop was at the dimer interface and surface exposed. (C) The X-ray crystal structure of the unbound GII.12 P domain monomer determined to 1.6-Å resolution (shown here as a modeled dimer) was colored according to monomers and P1 and P2 subdomains, i.e., chain A P1 (hot pink), chain A P2 (orange), chain B P1 (green), and chain B P2 (cyan). As for the GII.10 P1-interface loop, the GII.12 P1-interface loop was at the dimer interface and surface exposed. (D) Superposition of GII.10 (PDB ID, 3ONU), GII.12 (PDB ID, 3R6J), GII.4 (PDB ID, 2OBR), and GI.1 (PDB ID, 2ZL5), colored as shown in panels B and C, dark gray, and light gray, respectively, indicated that the four structures were very similar, except for several differences, including the P1-interface loop and the P2-extended loop. (E) The P1-interface loop (a close-up 90° rotation of panel D) was located at a dimer interface for all four structures. The lengths of the P1-interface loops were the same for GII.10, GII.12, and GII.4 but shorter for GI.1 (residues 445 to 456, 432 to 443, 436 to 447, and 425 to 431, respectively).