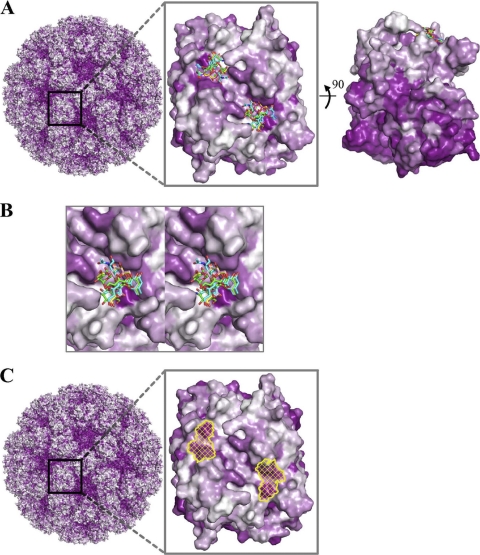
Fig. 8.
Surface representations of GII amino acid conservation and putative site of vulnerability for GII noroviruses. Antigenic diversity of noroviruses is seen primarily on the outermost surface of the capsid, although patches of conservation on the top surface are observed. The most prominent of these patches correspond to the P domain-binding sites of the HBGAs described here. (A) An alignment of GII genotypes was used to map the amino acid conservation and variability on the GII.10 P domain dimer structure. The color-coded conservation ranged from a deep purple, represented by highly conserved amino acids, to white, represented by highly variable amino acids. GII conservation was mapped onto a model of the viral capsid (left), with a zoomed-in P domain dimer outer-facing surface (middle) and a 90° dimer rotation that shows the difference in conservation of the outer- and inner-facing surfaces (right). The outer-facing surface (top portion) is substantially less conserved, with two major surface patches of conserved residues overlapping the HBGA binding site. (The highly conserved but nonprotruding portions of the capsid correspond to the shell domain.) (B) Close-up stereo view of panel A, middle, showing the six different HBGAs bound to the GII.10 P domain. (C) Surface representation of GII.10 amino acid conservation was obtained as described above and mapped onto the GII.10 P domain structure. The identified site of vulnerability (yellow) was defined as the surface area of the following GII.10 residues participating in conserved hydrogen-bonding interactions with αfucose1-2: Asn355, Arg356, Asp385 from one subunit, and Gly451 from the other subunit.