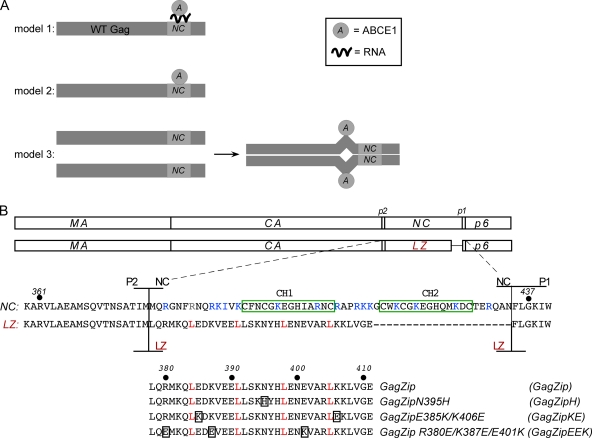
Fig. 1.
Model for ABCE1 binding and amino acid alignments of GagZip constructs. (A) Three models for the binding of ABCE1 to Gag. (Model 1) The binding of ABCE1 to Gag could be mediated through RNA recruited by NC. (Model 2) ABCE1 could bind directly to NC through protein-protein interactions. (Model 3) NC-mediated dimerization of Gag could expose a cryptic ABCE1 binding site in another region of Gag. (B) Amino acid alignments of Gag, GagZip, and GagZip mutants. Line diagrams show the domains of WT Gag (matrix [MA], capsid [CA], nucleocapsid [NC], and p6, along with two spacers, p2 and p1) and the GagZip chimeric protein used in this study. The sequences below show the amino acids in and around NC and the leucine zipper (LZ). The two zinc fingers in NC are boxed in green; basic residues in NC are indicated in blue; and amino acids in the d position of the LZ heptad repeat are indicated in red (see Fig. S1 in the supplemental material for more details). The alignment at the bottom shows the leucine zipper sequences of GagZip and the GagZip mutants, with mutated residues boxed.