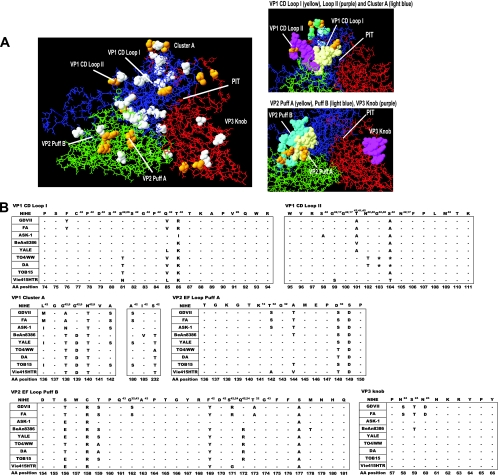
Fig. 7.
Analysis of the NIHE capsid structure formed by VP1, VP2, and VP3 proteins. Capsid proteins (VP1, VP2, and VP3) were aligned and overlaid onto the three-dimensional structure of the BeAn8386 strain using DeepView/Swiss-pdbViewer (version 4.0.1). (A) Quaternary structure with the individual proteins VP1 (blue), VP2 (green), and VP3 (red) indicated. White space-filled molecules indicate amino acid positions that differ between the sequence of BeAn8386 and NIHE. Amino acid positions that are unique to NIHE (compared to all TMEV strains) are highlighted in yellow. Highly variable regions, loops, puffs, and a canyon or “pit” are annotated. Also shown, to the right, are a close-up of VP1 CD loops I and II and the cluster A region (upper), as well as VP2 puff A and B and the VP3 knob (lower). (B) Amino acid alignments for the indicated region of all TMEV strains compared to the NIHE sequence. Dashes indicate identity to the NIHE sequence, and the single-letter codes for nonidentical amino acids are shown. Amino acid positions and annotations are the same as those described in the legend to Fig. 6.