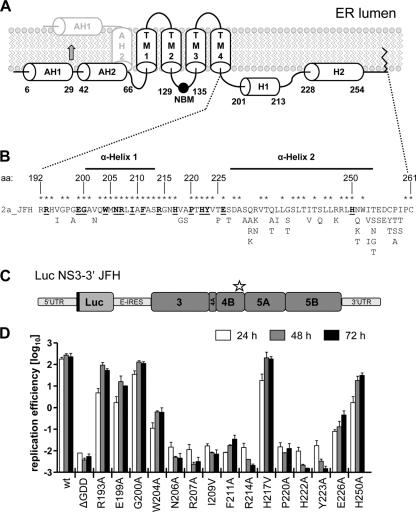
Fig. 1.
Residues in the C-terminal domain of NS4B are important for HCV RNA replication. (A) Predicted NS4B membrane topology. The schematic representation shows the N-terminal amphipathic α-helices AH1 and AH2, the four transmembrane segments (TM1 to TM4), the nucleotide binding motif (NBM), the C-terminal α-helices H1 and H2, and the very C-terminal palmitoylation site(s). The reported posttranslational flip of the N terminus into the ER lumen is indicated by the arrow. (B) HCV JFH1 protein sequence (2a_JFH) of the NS4B C-terminal region. NS4B amino acid (aa) positions are given. Fully conserved amino acids are marked with asterisks. Alternative amino acid residues found in other genotypes are indicated below. Residues targeted in this mutagenesis study are underlined and highlighted with bold letters. (C) Schematic representation of the subgenomic luciferase reporter replicon. The luciferase is expressed as an N-terminal fusion with 16 aa of the N-terminal region of the core protein (black line) and is translated under the control of the HCV IRES contained in the 5′UTR. The second cistron (NS3 to NS5B) is translated via the internal ribosome entry site of the encephalomyocarditis virus (E-IRES). The star indicates the region in NS4B targeted by the mutations. (D) Huh7 cells were transfected with in vitro transcribed luciferase replicon RNAs specified in the bottom. Cells were lysed 4, 24, 48, and 72 h after transfection, and the luciferase activity in cell lysates was determined. The data were normalized to the 4-h value that reflects transfection efficiency. The background of the assay is determined with the active-site NS5B polymerase mutant (ΔGDD). The mean values of two independent experiments are shown. Error bars indicate standard deviations.