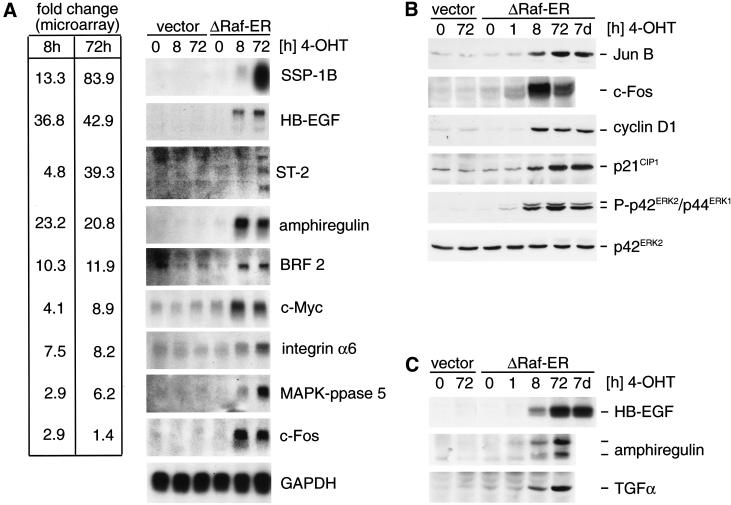
Figure 3.
Differential expression of Raf target genes. (A) Northern blot detection of Raf target genes identified by microarray analysis. RNA from empty vector or ΔRaf-ER MCF-10A cells treated with 4-OHT for the indicated times while cultured in minimal medium for 24 h before lysis (in the same way as described for the experiment shown in Table 1) was separated by denaturing electrophoresis, blotted onto nylon membranes, and probed with radioactively labeled cDNA probes for the indicated genes. For comparison, fold change values measured in the microarray experiment (Table 1) are indicated on the left. (B) Whole cells lysates from cells prepared in parallel to samples used for RNA preparation in 3A were used to detect JunB, c-Fos, cyclin D1, and p21CIP1 by immunoblotting. In addition, whole cell lysates from MCF-10A ΔRaf-ER cells treated with 4-OHT for 7 d were used (lane 7). Activation of ΔRaf-ER by 4-OHT was monitored by detection of p42ERK2/p44ERK1 MAPK phosphorylation using a phosphospecific antibody. (C) Same cell lysates as in B were used to detect HB-EGF, TGFα, and amphiregulin by immunoblotting. Amphiregulin was detected as two bands representing the 17-kD precursor and a smaller processed form.