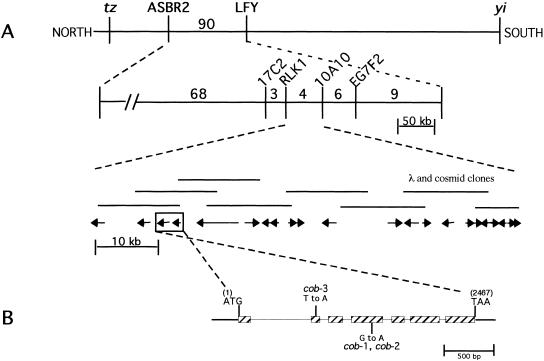
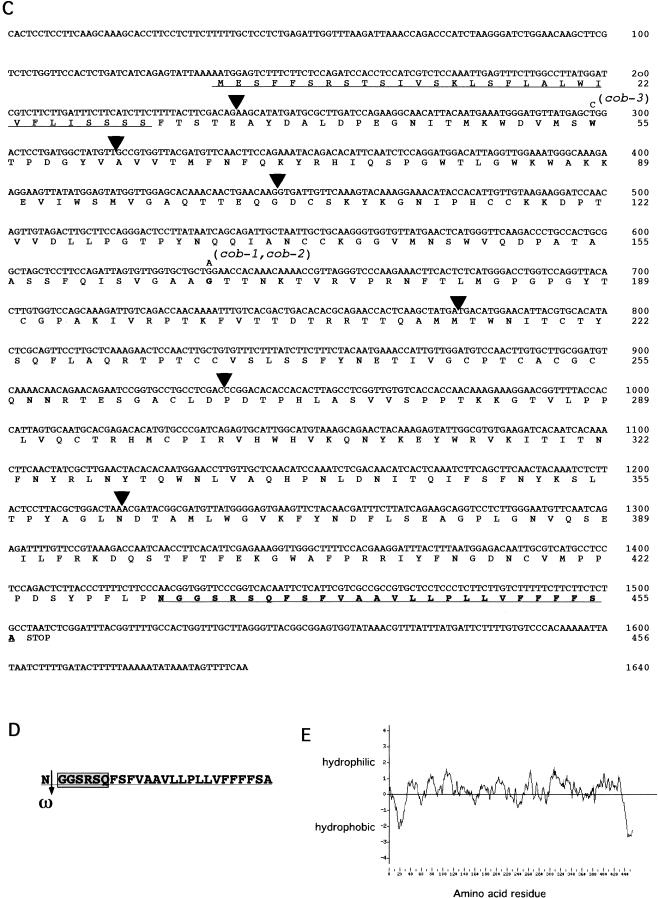
Figure 3.
Cloning of COB and analysis of its deduced amino acid sequence. (A,B) Summary of the chromosome walk. (A) Initial mapping placed COB near the LFY locus. Three point crosses using yi tz and cob-1 plants identified crossover events between the CAPS markers ASBR2 and LFY (vertical lines). Analysis of internal CAPS markers placed COB in the 74 kb interval between the RLK1 and the 10A10 CAPS markers. (Numbers represent the number of recombinants isolated between the markers shown). The 22 putative genes or open reading frames in this region were sequenced from the cob-1 background until a mutation was revealed in a putative gene (shown in the boxed region; arrows indicate the direction of transcription). (B) Genomic organization of COB (hatched boxes represent exons). Positions of the translational start (ATG) and stop (TAA) of the predicted coding sequence are shown. The position and nucleotide changes in the mutant alleles are also shown. (C) The predicted amino acid sequence is shown directly below the cDNA sequence. Numbers to the right refer to the positions of nucleotides or amino acid residues. Triangles represent the positions of introns. Missense point mutations in cob-1, cob-2, and cob-3 alleles are shown above the nucleotide sequence. Underlined is the putative cleavable N-terminal signal sequence (as determined with pSORT [http://psort.nibb.ac.jp/]). Underlined and in bold is the motif, which in addition to the N-terminal signal sequence and hydrophilic central portion, appears to meet all of the sequence requirements for processing and GPI linkage and the GPI prediction site (http://mendel.imp. univie.ac.at/). In bold are the amino acids, which would be changed in the mutant alleles. (D) The GPI linkage motif in COB includes the predicted cleavage (ω) site, which is the residue to the left of the cleavage (arrow). ω +1 and ω +2 sites immediately following the cleavage site, which are part of a six amino acid spacer region (boxed), and a 19 amino acid hydrophobic tail (see text for consensus residues). (E) Hydropathy analysis of COB shows the hydrophobic N- and C-terminals and the hydrophilic central portion.