Figure 3.
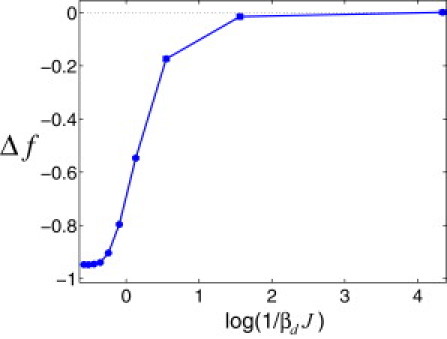
Average TF-DNA binding free energy, , numerically computed at different values of the design temperature, where , with the partition function for an entirely random DNA sequence. We designed 200 sequences with length at each Td. We performed 5 × 106 MC steps to design each sequence, attempting in each step to exchange two basepairs chosen at random. The overall nucleotide composition for each sequence was uniform and fixed. The design potential was (attraction) for identical nearest-neighbor basepairs and (repulsion) for different nearest-neighbor basepairs, with . The contact energies, , were drawn from a Gaussian distribution, , with zero mean, , and standard deviation for each nucleotide type α. We computed as an average over 250 TFs and 200 sequences at each Td and used . The error bars are calculated as specified in Fig. 2, and they are smaller than the marker size.
