Abstract
5S rRNA is an essential component of ribosomes of all living organisms, the only known exceptions being mitochondrial ribosomes of fungi, animals, and some protists. An intriguing situation distinguishes mammalian cells: Although the mitochondrial genome contains no 5S rRNA genes, abundant import of the nuclear DNA-encoded 5S rRNA into mitochondria was reported. Neither the detailed mechanism of this pathway nor its rationale was clarified to date. In this study, we describe an elegant molecular conveyor composed of a previously identified human 5S rRNA import factor, rhodanese, and mitochondrial ribosomal protein L18, thanks to which 5S rRNA molecules can be specifically withdrawn from the cytosolic pool and redirected to mitochondria, bypassing the classic nucleolar reimport pathway. Inside mitochondria, the cytosolic 5S rRNA is shown to be associated with mitochondrial ribosomes.
Keywords: MRP-L18, mitochondria, human, ribosome, RNA import, chaperone
Among ribosomal RNA molecules, 5S rRNA occupies a very particular place. This only ∼120-nucleotide (nt)-long nucleic acid displays an unusually complex structure with outstanding protein interaction capacities, essential for both its intracellular transport and function inside ribosomes (for review, see Szymanski et al. 2003; Smirnov et al. 2008b). One of the best conserved ribosomal macromolecules, 5S rRNA, has been shown to perform the function of the higher regulatory center, providing the cross-talk among almost all functional sites of the translating ribosomal machine (Bogdanov et al. 1995; Smith et al. 2001; Kiparisov et al. 2005; Kouvela et al. 2007). The major 5S rRNA-binding protein of the ribosome is prokaryotic protein L18 and its eukaryotic homolog, eL5. Proteins of this family, named eL5/L18, are present in all living systems and are absolutely needed for 5S rRNA integration into the ribosomal large subunit (DiNitto and Huber 2003). In bacterial ribosomes, 5S rRNA also interacts with proteins L5 (despite its name, it is not a homolog of the eukaryotic eL5) and L25, thus forming a main part of the central protuberance of the large subunit (Ban et al. 2000). The recently reported crystal structure of the yeast Saccharomyces cerevisiae 80S ribosome demonstrates tight interaction of 5S rRNA with protein eL5, as well as with eL11 (homolog of prokaryotic L5, found only in yeast ribosomes) and eL6. The central protuberance of the large subunit, dominated by 5S rRNA, undergoes considerable structural rearrangement upon mRNA translocation and may coordinate changes in different sites of the ribosome (Ben-Shem et al. 2010).
Notwithstanding its important role, attempts to detect 5S rRNA in mitochondrial ribosomes of fungi, animals, and some protists have been unsuccessful (Koc et al. 2001b; Sharma et al. 2003, 2009). In the yeast S. cerevisiae and the kinetoplastid protozoan Leishmania tarentolae, 5S rRNA is not encoded by the mitochondrial genome, and there is no evidence that this RNA can be imported from the cytosol. Although, in such dynamically evolving mitochondrial systems as that of L. tarentolae, recruitment of novel ribosomal proteins could have ensured the functional complementation for the lost ancestral 5S rRNA (Smits et al. 2007; Sharma et al. 2009), it appears unlikely in the case of mammalian mitochondria, where the additional and enlarged ribosomal proteins do not compensate for the majority of the missing rRNA segments (Sharma et al. 2003).
The mammalian mitochondrial genome encodes for 13 proteins, 22 tRNAs, and two rRNAs, the latter being components of small 28S and large 39S subunits of the 55S mitochondrial ribosome (O'Brien 2002). Thus far, RNA components encoded by the mitochondrial genome are believed to be sufficient for mitochondrial translation of the 13 structural genes coding for inner membrane proteins essential for oxidative phosphorylation (Attardi and Ojala 1971). An intriguing discovery that a significant portion of nuclear DNA-encoded 5S rRNA is naturally moved into mammalian mitochondria (Yoshionari et al. 1994; Magalhaes et al. 1998; Entelis et al. 2001) raised questions about both the molecular mechanism of this targeting and its biological significance. In our previous study, we identified structural elements of 5S rRNA serving as signals of its mitochondrial import, localized in the proximal part of helix I and in the loop D–helix IV region (Fig. 1A; Smirnov et al. 2008a). It had also been hypothesized that these regions correspond to binding sites with protein factors recruited to transport the RNA to the organelles. One of these factors was identified as the mitochondrial enzyme rhodanese (thiosulfate sulfurtransferase), interacting with both α-domain and γ-domain regions of 5S rRNA, but binding mainly to the proximal part of helix I (Smirnov et al. 2010). It has also been shown that 5S rRNA import is critical for mitochondrial translation, suggesting that this molecule might have a closer association with the organellar genetic system expression than it had been traditionally believed (Smirnov et al. 2010).
Figure 1.
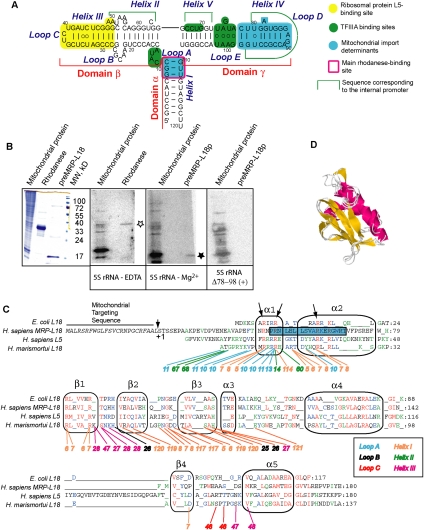
Mitochondrial ribosomal protein L18, a member of the L18/eL5 family. (A) The secondary structure of human 5S rRNA and its main functional sites (Smirnov et al. 2008a,b, 2010). (B) North–Western analysis of HepG2 mitochondrial proteins for binding to human 5S rRNA. (Left) Coomassie staining of mitochondrial or recombinant proteins. (Panels 2–4) Autoradiograph of proteins transferred to nitrocellulose membrane after incubation with labeled human 5S rRNA, folded in the absence (to adopt a branched conformation) or presence of Mg2+, or containing deletion in the γ-domain (as indicated below the panels). The asterisks indicate the band corresponding to rhodanese (empty) or MRP-L18 (solid). The last filter hybridized with the Δ78–98 5S rRNA version was overexposed in comparison with the first two [as indicated by (+)]. (C) Alignment of sequences of four members of the L18/eL5 family of ribosomal proteins from different kingdoms: E. coli L18 (bacteria), Homo sapiens MRP-L18 (mammalian mitochondria), H. sapiens L5 (mammalian cytosol), and Haloarcula marismortui L18 (archaea). Alignment was done according to Koc et al. (2001b) and Woestenenk et al. (2002) and corrected manually. Secondary structure elements are mapped according to H. marismortui and Thermus thermophilus L18 structures. Universally conserved positions are in red, those shared by three out of four sequences are in blue, and those shared by two out of four sequences are in green. The preMRP-L18 presequence is given in black italic. The N-terminal helix predicted by Psipred (Bryson et al. 2005) and Porter (Pollastri and McLysaght 2005) software for MRP-L18 is framed in a blue box. Numbers below sequences represent 5S rRNA nucleotides implicated in interactions with corresponding H. marismortui L18 protein residues. Conserved arginine residues in the N-terminal part of the protein are indicated by arrows. (D) Superposition of three structural models developed by Geno3D2 (Combet et al. 2002) for MRP-L18 (residues 56–170) and ribosomal protein L18 of Th. thermophilus (residues 25–115).
In this study, we identify the second protein factor involved in both targeting of 5S rRNA and its intramitochondrial function. We show that human mitochondrial ribosomal protein L-18 (MRP-L18), a homolog of the Escherichia coli large ribosomal subunit protein L18, a member of a protein family L18/eL5 embracing the most conserved 5S rRNA-binding ribosomal proteins (Ban et al. 2000; DiNitto and Huber 2001), has been adapted to bind human cytosolic 5S rRNAs. In cytosol, this binding can induce the conformational change needed for the 5S rRNA to be recognized by rhodanese. Thus, rhodanese and the cytosolic precursor of MRP-L18 function in a molecular conveyor, allowing an efficient uptake of cytosolic 5S rRNA molecules and their redirecting into mitochondria. Imported into mitochondria and processed, mature MRP-L18 retains the capacity of interaction with the imported 5S rRNA. Finally, for the first time, evidence has been obtained that the cytosolic 5S rRNA is associated with the mammalian mitochondrial ribosome, which can be a new adaptive mechanism in cytoplasm–mitochondria cross-talk.
Results
Mitochondrial ribosomal protein L18 binds the cytosolic 5S rRNA
In our previous work, we have shown that, for its import into human mitochondria, 5S rRNA needs at least two soluble protein factors, one of which was identified as rhodanese (Smirnov et al. 2010). Interestingly, rhodanese was found to bind to 5S rRNA only if the latter is folded into an unusual “branched” conformation, suggesting that an unknown upstream factor with an RNA chaperone activity should be implicated. Having expected its association with mitochondria, we analyzed the total mitochondrial protein extract from HepG2 human cells for the presence of 5S rRNA-binding proteins using North–Western analysis (Fig. 1B). Several proteins were found to possess the 5S rRNA-binding activity. One of them (∼33 kDa) might correspond to rhodanese (Fig. 1B, panel 2). However, the most prominent signal of interaction with labeled human 5S rRNA was detected for a set of proteins migrating in the area of 18–20 kDa. This gel area is extremely heterogeneous, and more than one component may be responsible for the signal observed in North–Western experiments (indeed, a huge majority of mitoribosomal proteins migrate in this area, especially in such a loose gel). Among mitochondrial proteins of this size, one of the most obvious candidates was the ribosomal protein MRP-L18. MRP-L18 was chosen for further detailed investigation mainly since, among all mitochondrial proteins of this size, it belongs to the family of the best conserved 5S rRNA interactors. To test this possibility, we purified recombinant MRP-L18 and demonstrated that it migrates in the same 18- to 20-kDa area of SDS-PAAG and can interact with labeled human 5S rRNA in the conditions used for North–Western analysis (Fig. 1B, panel 3). Moreover, detected interaction was not due to nonspecific RNA-binding properties shared by most of ribosomal proteins, since a mutant version of 5S rRNA containing a deletion in the γ-domain (Fig. 1A; Table 1) has lost the ability to bind recombinant MRP-L18 that also correlated with a visible decrease of interaction with 18- to 20-kDa mitochondrial proteins (Fig. 1B, panel 4). A band with a similar size was still detected in mitochondrial lysate; so far, since this is a complex mixture of proteins (see above), one cannot be affirmative concerning the identity of the protein(s) giving the signal in this case. Nevertheless, these data indicate that the ribosomal protein MRP-L18 might be at least one of the human mitochondrial proteins detected on our preparation capable of binding cytosolic 5S rRNA.
Table 1.
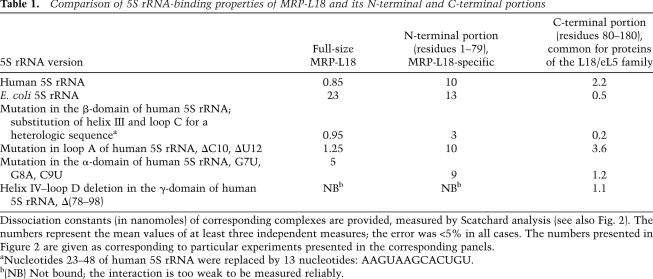
Comparison of 5S rRNA-binding properties of MRP-L18 and its N-terminal and C-terminal portions
Identified recently as a component of the large subunit of mammalian mitochondrial ribosome, MRP-L18 was described as a homolog of prokaryotic protein L18, a member of the L18/eL5 family, on the basis of a sequence alignment (Koc et al. 2001b; Rorbach et al. 2008). The current proteome analysis of mammalian mitochondrial ribosome suggests that the mitochondrial homologs of other 5S rRNA-binding proteins (L5 and L25) are missing (Sharma et al. 2003). Since some doubts were harbored about the putative affinity of MRP-L18 to 5S rRNA (Koc et al. 2001b), we decided to readdress this question in a more detailed way.
Prediction of the tertiary structure of MRP-L18 by Geno3D2 (Combet et al. 2002) and 3D-JIGSAW (Contreras-Moreira and Bates 2002) software reveals conservation of the structural core characteristic of the family: a four-stranded anti-parallel β-sheet and a bundle of three α-helices (Fig. 1C,D). Since this new alignment brings together four representatives of major domains of life that have certainly diverged upon evolution, we observe the appearance of multiple gaps. For comparison, when very similar animal MRP-L18s were aligned against E. coli L18 only (Koc et al. 2001b), the number of gaps was quite low, indicating a much closer relationship between bacterial and mitochondrial representatives of the family. Another important reason for gapping was the use of a structure-based approach, meaning that not only were sequence similarities taken into consideration, but also secondary structure elements’ distribution was respected. For this, we used structural information available for bacterial and archaeal L18s and secondary and tertiary structure predictions for MRP-L18. After realignment of sequences, the conserved arginine residues of the N-terminal part of the protein needed for the interaction with 5S rRNA (Furumoto et al. 2000) and ignored in previous work (Koc et al. 2001b) were also detected (Fig. 1C).
To directly address the RNA-binding properties of MRP-L18, gel shift assays followed by Scatchard plot analyses were performed (Fig. 2; Table 1). Surprisingly, MRP-L18, as well as its precursor form, preMRP-L18 (data not shown), was able to interact with both human (eukaryotic type) and E. coli (bacterial type) 5S rRNAs, although with different affinities—the human 5S rRNA appearing to be a better ligand (Fig. 2A; Table 1). In order to localize MRP-L18-binding sites on human 5S rRNA, we measured dissociation constants of complexes between MRP-L18 and several mutated versions of human 5S rRNA (Table 1). These mutations, although not identical to those described in other studies, were created to disrupt, exchange, or delete the structural elements previously shown to be involved in L18 or eL5 binding in both structural (Ban et al. 2000; Woestenenk et al. 2002) and biochemical (Aoyama et al. 1984; Huber and Wool 1984; Scripture and Huber 1995; Gongadze et al. 2001) studies. Mutations in the β-domain and loop A—sites normally responsible for interaction of 5S rRNA with L18/eL5 proteins (Ban et al. 2000)—did not significantly influence the binding. In contrast, the helix IV–loop D deletion in the γ-domain of human 5S rRNA drastically inhibited interaction (Figs. 1B, 2A), indicating that this element, identified previously as one of the 5S rRNA mitochondrial import determinants (Fig. 1A), represents the binding platform for MRP-L18.
Figure 2.
5S rRNA-binding properties of MRP-L18. (A) Scatchard plot determination of dissociation constants for complexes between MRP-L18 and 5S rRNA versions. Human 5S rRNA mutants used were mutation in loop A (ΔC10 and ΔU12), mutation in the α-domain (G7U, G8A, and C9U), mutation in the β-domain (a substitution of helix III and loop C for a heterologous sequence) (Table 1), and mutation in the γ-domain [Δ(78–98)]. The complex is labeled with an arrow. (B,C) An example of Scatchard plot determination of dissociation constants for complexes between the N-terminal (residues 1–79) (left panel) or C-terminal (residues 80–180) (right panel) portions of MRP-L18 and human 5S rRNA versions G7U, G8A, C9U, and Δ(78–98) (see also Table 1). One micromolar and 40 nM purified protein were used, respectively.
Unlike other members of the eL5/L18 family, the N-terminal domain of MRP-L18 is mostly disordered (as predicted by Psipred [Bryson et al. 2005] and Porter [Pollastri and McLysaght 2005] software) and contains only one long α-helix (Fig. 1C). These particular features encouraged us to compare 5S rRNA-binding properties of the C-terminal portion of the protein (residues 80–180) with those of the N-terminal part (residues 1–79). It appeared that both of them have affinity to 5S rRNA (Fig. 2B,C; Table 1), but a clear distribution of functions between these two parts of the protein was evident. Whereas the C-terminal part, inherited from a bacterial ancestor, provides a rather strong affinity to all types of 5S rRNA (even the γ-domain mutant is bound by it) (Table 1), the mitochondria-specific N-terminal extension of MRP-L18 seems to be responsible for discrimination between mutant 5S rRNA versions, thus conferring to the whole protein the ability to bind the cytosolic 5S rRNA in a noncanonical way. One cannot, in principle, exclude the possibility that the C terminus's behavior may be a consequence of contamination. Indeed, the preparation of this protein was not absolutely free of copurifying proteins (although it was very highly enriched). Still, differences in Kd observed for various 5S rRNA versions (Table 1) and the very range of these values, which are close to those observed for highly purified full-size MRP-L18 protein, permit us to neglect the possibility of nonspecific association with contaminants.
PreMRP-L18 and rhodanese form the minimal 5S rRNA mitochondrial import vehicle
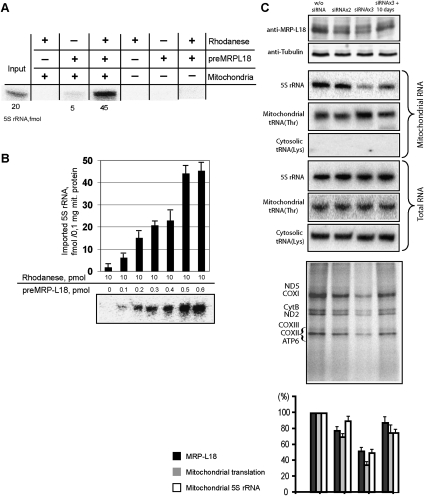
As demonstrated above, the binding site for MRP-L18 coincides with one of the 5S rRNA mitochondrial import determinants (Fig. 1A; Smirnov et al. 2008a). It raised the question of a possible involvement of preMRP-L18 in the process of 5S rRNA transport to mitochondria. To address it, the standard in vitro 5S rRNA import assay (Entelis et al. 2001) was used (Fig. 3A). Isolated human mitochondria were coincubated with radioactively labeled 5S rRNA in the presence of ATP and protein factors of interest. The efficiency of 5S rRNA uptake by mitochondria was evaluated by measuring the radioactive signal of the imported RNA, all nonimported molecules having been destroyed by RNase treatment. When taken separately, rhodanese and preMRP-L18 failed to direct 5S rRNA import. But, when the combination of both proteins was assayed, the level of 5S rRNA mitochondrial uptake was comparable with that obtained with crude protein preparation. Furthermore, the import efficiency was clearly dependent on the preMRP-L18 concentration (Fig. 3A,B). Thus, the minimum in vitro system of 5S rRNA import into human mitochondria was reconstituted by combining purified rhodanese and preMRP-L18. It should also be noted that, like rhodanese (Smirnov et al. 2010), preMRP-L18 does not need 5S rRNA for its own targeting into mitochondria (Supplemental Fig. S1).
Figure 3.
PreMRP-L18 as a 5S rRNA mitochondrial import factor. (A) Reconstitution of the minimum 5S rRNA import system in vitro. Example of in vitro mitochondrial import assay for labeled 5S rRNA in the presence of rhodanese (10 pmol) and preMRP-L18 (0.5 pmol); an autoradiograph of RNA protected from nuclease cleavage is presented. Rhodanese was used in its urea-denatured form as described in Smirnov et al. (2010). (B) The dose dependence of 5S rRNA import into isolated human mitochondria upon preMRP-L18 amount. (C) Effect of preMRP-L18 knockdown on 5S rRNA import into HepG2 mitochondria. Western blots of total cellular protein extracts, corresponding Northern blots of total and mitochondrial RNAs, and autoradiographs of SDS gels with mitochondrial translation products are provided. (W/o siRNA) Mock-transfected cells (showed no difference from ones transfected with siRNA against luciferase mRNA) (data not shown); (siRNAx2) cells transfected twice (spaced by 3 d) with MRP-L18-directed siRNAs (analyzed 3 d after the second transfection); (siRNAx3) cells transfected three times (spaced by 3 d) with MRP-L18-directed siRNAs (analyzed 3 d after the third transfection); (siRNAx3 + 10 d) recovery of MRP-L18 expression 10 d after the last transfection. Correlations between MRP-L18 expression, 5S rRNA mitochondrial import, and mitochondrial translation levels are presented in the histogram below the experimental panels. The values in mock-transfected cells were taken as 100%. The bars represent the range of values obtained in two independent experiments.
To further confirm preMRP-L18 as a 5S rRNA mitochondrial import factor in vivo, we studied the dependence between expression levels of the protein and the amount of 5S rRNA inside mitochondria in living human cells. For this, expression of preMRP-L18 was partially inhibited by transient transfection of HepG2 cells with specific siRNAs, and the effect of this inhibition on 5S rRNA import was studied (Fig. 3C). Although the efficiency of silencing was low, a good correlation between the intracellular level of preMRP-L18 and the mitochondrial level of 5S rRNA was observed. Decrease of preMRP-L18 expression led to a proportional fall of 5S rRNA import, while restoration of its expression in 10 d after the last siRNA transfection coincided with an equally proportional increase of 5S rRNA in mitochondria. Neither transcription of mitochondrial DNA nor the overall cellular 5S rRNA level was affected, indicating that the effect of the short-term protein's withdrawal on the 5S rRNA level inside mitochondria was specific. This result allows us to suggest that preMRP-L18 is indeed involved in 5S rRNA mitochondrial import and/or stabilization in vivo. As expected, the changes in the MRP-L18 and 5S rRNA mitochondrial levels were associated with a corresponding decrease/restoration of the general mitochondrial translation activity (Fig. 3C).
PreMRP-L18 and rhodanese function in a molecular chaperone conveyor to ensure mitochondrial 5S rRNA targeting
Rhodanese and preMRP-L18 interaction sites on 5S rRNA partially overlap. Indeed, we have shown previously that deletion of the γ-domain import determinant leads to a >200-fold increase in the Kd of the rhodanese–5S rRNA complex. This is why we suggested that this region functions as an additional binding platform for rhodanese, although the α-domain site remains critical for the interaction (Smirnov et al. 2010). At the same time, the γ-domain also serves as the preMRP-L18-binding platform (Fig. 1A). Therefore, two questions arise: (1) How do the two precursor proteins bind 5S rRNA prior its import into mitochondria? (2) How do their mature forms interact with 5S rRNA inside mitochondria?
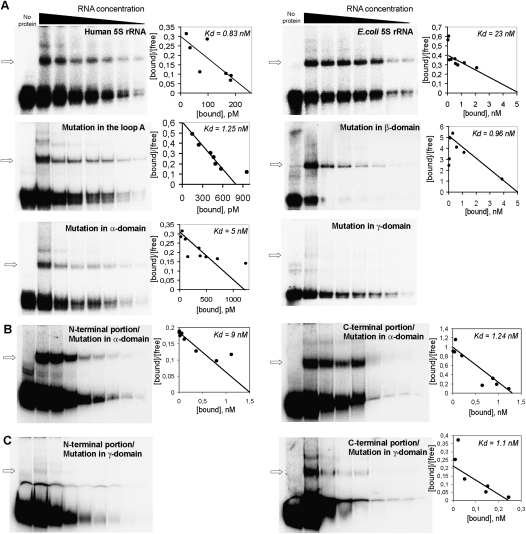
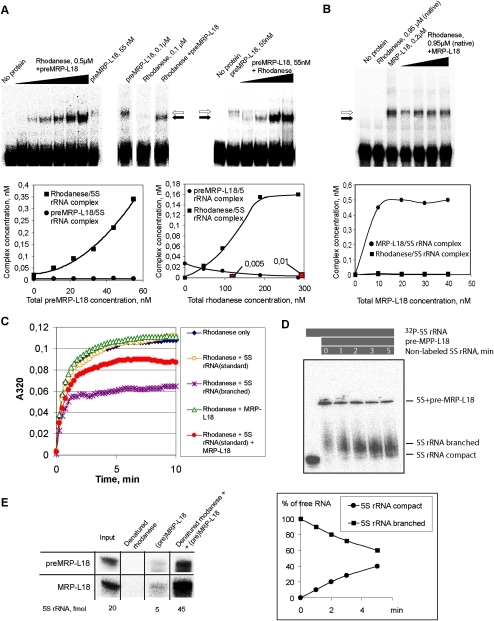
To address these questions, we studied 5S rRNA behavior in the two-protein system by the mobility shift approach (Fig. 4A,B). Two situations were examined. In the first one, human 5S rRNA was incubated with both proteins in their precursor forms (preMRP-L18 and misfolded rhodanese) (Smirnov et al. 2010). With the increase of rhodanese concentration (Fig. 4A, right panel), its complex with 5S rRNA was sharply increased until saturation, whereas the complex with preMRP-L18 suffered a gradual decline. Noteworthy, in the presence of preMRP-L18, the rhodanese/5S rRNA complex was formed with a more than an order higher efficiency than with rhodanese alone (Fig. 4A, middle panel). But the most striking feature was that, with the increase of preMRP-L18 concentration, the 5S rRNA–preMRP-L18 complex was not increased, but, rather, the 5S rRNA–rhodanese one was, gradually appearing as more prominent (Fig. 4A, left panel). Such a pattern is characteristic for consecutive reactions, indicating that the first protein (preMRP-L18) binds to the substrate (5S rRNA) and then transfers it to the second protein (rhodanese). No ternary complex between the RNA and both proteins could be observed, which means that this intermediate state, if any, is short-lived, while the transfer rate is high. These results enabled us to elaborate a kinetic model revealing the sequential two-step mechanism of 5S rRNA transfer (see Supplemental Fig. S2; Supplemental Material for details). The preMRP-L18/rhodanese system appears to function as a very fast conveyor, which can ensure an efficient uptake of 5S rRNA molecules by preMRP-L18 (k1 ≈ 4 × 105 M−1 sec−1, 25°C), followed by a rapid discharge on the recipient protein, rhodanese (k2 ≈ 5 × 107 M−1 sec−1, 25°C).
Figure 4.
The conveyor mechanism of 5S rRNA binding by preMRP-L18 and rhodanese. (A) Mobility shift assays of complexes formed by 5S rRNA in the presence of increasing amounts of urea-denatured rhodanese (right panel) or preMRP-L18 (left panel) (with the constant concentration of the second protein). Complexes with rhodanese and preMRP-L18 are marked with black and white arrows, respectively. Rhodanese is present in all samples except “No protein” and “preMRP-L18.” Concentration dependences for both complexes are shown below the corresponding autographs. Red columns correspond to the rhodanese/5S rRNA complex formed in the absence of preMRP-L18. (B) Mobility shift assays of complexes formed by 5S rRNA in the presence of both native rhodanese and increasing amounts of mature MRP-L18. Designations are the same as in A. (C) Rhodanese aggregation assays in the presence of 3 μM 5S rRNA in either the standard or the branched conformation (Smirnov et al. 2010), 3 pM preMRP-L18, or 3 μM 5S rRNA in the standard conformation preincubated for 5 min with 3 pM preMRP-L18. (D) Release of labeled 5S rRNA from its complex with pre-MRP-L18 in the presence of excess nonlabeled competitor 5S rRNA. The presence of the labeled RNA and protein and nonlabeled competitor are indicated by gray bars above the panel, time of incubation is indicated inside the bottom bar, and the different forms of 5S rRNA are indicated at the right. A quantification graph is shown below the autoradiograph. (E) Comparison of minimum in vitro 5S rRNA import systems composed of urea-denatured rhodanese (10 pmol) and 0.5 pmol of either preMRP-L18 (top line) or mature MRP-L18 (bottom line). Autoradiograph of imported (protected from nuclease cleavage) 5S rRNA is presented, as in Figure 3A.
The situation described for cytosolic forms of both 5S rRNA import factors differs drastically from what was observed when this RNA was found in the presence of their mitochondrial forms (Fig. 4B). In this latter case, no 5S rRNA–rhodanese complex was formed as a consequence of the loss by rhodanese of RNA-binding activity upon folding into its enzymatic conformation, as was shown previously (Smirnov et al. 2010). On the other hand, MRP-L18 has the same affinity to 5S rRNA as its precursor, and thus it remains the mitochondrial protein capable of interacting with the imported 5S rRNA in the organelle. These data suggest the existence of an elegant molecular mechanism, which implies an interplay between (pre)MRP-L18 and rhodanese during and after 5S rRNA transport into mitochondria. It is clear that additional experimentation is needed to definitely validate this model; so far, all results described above, as well in vivo experiments hereafter, strictly corroborate it.
PreMRP-L18 function is 5S rRNA folding into import-competent conformation
As was shown previously, only a particular “branched” conformation of human 5S rRNA is appropriate for chaperone-like interaction with misfolded rhodanese (Smirnov et al. 2010). We hypothesized that induction of such a conformation could be a specific function of the second import factor. Indeed, preMRP-L18 seems to be well equipped to complete this function, since proteins of the eL5/L18 family are known to have RNA chaperone activity in bacterial, archaean, and eukaryotic cells (Pace et al. 1984; DiNitto and Huber 2003; Semrad et al. 2004; Scripture and Huber 2011), and the distribution of intrinsic disorder regions in preMRP-L18 essential for performing such a task (Tompa and Csermely 2004; Kovacs et al. 2009) closely follows that in bacterial L18 (Supplemental Fig. S3).
Our data show that preMRP-L18 ensures a much more efficient binding of 5S rRNA by rhodanese (Fig. 4A). Therefore, we hypothesized that preMRP-L18 might bind to 5S rRNA and confers to it a more open conformation recognized by misfolded rhodanese. Thus, two consecutive chaperoning events may take place: 5S rRNA folded by preMRP-L18 then becomes a chaperone for misfolded rhodanese. We modeled this situation in the rhodanese aggregation assay (Fig. 4C). Such an approach was used since apparent dissociation constants, as measured by standard titration curves, have no significant difference between the two 5S rRNA forms, meaning that electrophoretic mobility shift assay (EMSA) does not reveal any preference for either form. Still, our previously described aggregation assay (Smirnov et al. 2010) indicates that there is an important functional difference between complexes formed in each case, with only 5S rRNAEDTA being protective against aggregation and showing cochaperone-like behavior. Thus, we showed that the aggregation suppression does depend on interaction between 5S rRNA and rhodanese, because 5S rRNAEDTA molecules mutated in any of the rhodanese-binding sites failed to prevent the protein from aggregation (Smirnov et al. 2010). Therefore, in this particular situation, the aggregation assay was considered more informative. Indeed, of two 5S rRNA conformations—the standard and the branched—only the second can suppress aggregation of urea-denatured rhodanese (Fig. 4C). Still, if the 5S rRNA in its standard conformation is briefly preincubated with catalytic amounts of preMRP-L18, each of them being inactive when taken separately, rhodanese aggregation becomes partially suppressed and even tends to be slightly reversed after 7 min of incubation, suggesting that the chaperone-active 5S rRNA form has been generated in this system.
To obtain further experimental evidence of the chaperone-induced conformational changes of 5S rRNA upon interaction with preMRP-L18, we developed an EMSA specially adapted for this purpose. Our earlier experiments showed that the branched conformation of human 5S rRNA is very unstable in solution (even if EDTA is used to remove divalent cations), and within minutes relaxes into the classic, compact form. Still, as we showed previously, it was possible to detect the branched form if rapid loading on a 10% Mg-free native gel was used (Smirnov et al. 2010). Here, we monitor the situation when complexes formed between Mg-folded 5S rRNA and MRP-L18 in a Mg-free buffer were challenged by an excess of unlabeled 5S rRNAMg (Fig. 4D). This is a routine test in RNA chaperone studies (for example, Chaulk et al. 2011). The exchange thus initiated resulted in progressive liberation of labeled 5S rRNA, and its mobility in native gel was slower than that of 5S rRNAMg. Moreover, accumulation of the compact conformation becomes evident in a few minutes of the chase, indicating that, even in a Mg-free buffer, this branched conformation very rapidly relaxes into the compact one. This experiment provides a direct proof of MRP-L18 RNA chaperone activity.
We reasoned also that, if the preMRP-L18’s only function is folding 5S rRNA into an import-competent conformation recognized by rhodanese, then the mature protein, which cannot enter mitochondria but has the same 5S rRNA-binding activity, should be as active in the mitochondrial import assay as its precursor. Indeed, the minimum import system composed of rhodanese and processed mature MRP-L18 provided internalization of 5S rRNA by isolated mitochondria with the same efficiency as the one composed of rhodanese and preMRP-L18 described above (Fig. 4E). These data prove that preMRP-L18 plays a mediator role in the 5S rRNA mitochondrial import pathway, while rhodanese functions as the carrier protein.
MRP-L18 interacts with 5S rRNA in vivo
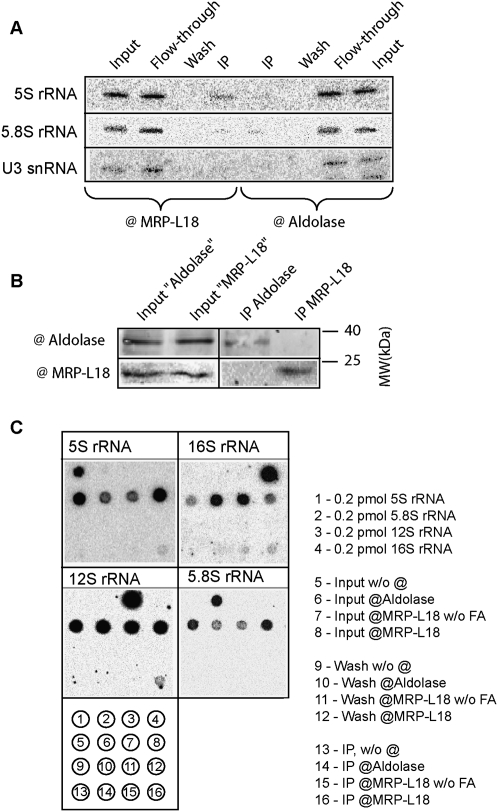
To confirm the interaction of 5S rRNA and MRP-L18 protein in vivo, we performed the cross-linking immunoprecipitation (CLIP) assay. To this end, HepG2 cells were submitted to formaldehyde treatment and lysed in conditions permitting the solubilization of mitochondrial material, and the RNA content was analyzed in immunoprecipitates generated by anti-MRP-L18 antibodies or—as control of nonspecific binding—anti-aldolase ones (Fig. 5). It appears that anti-MRP-L18-generated immunoprecipitates do contain detectable and reproducible amounts of 5S rRNA, which are not detected in anti-aldolase-generated immunoprecipitates (Fig. 5A). Furthermore, the same precipitates contain only trace amounts of contaminant nuclear 5.8S rRNA, and no difference for these trace amounts was observed in two immunoprecipitates. Finally, no U3 snRNA was detected in either immunoprecipitate. The efficiency of the test was monitored by Western analysis (Fig. 5B). Indeed, for both immunoprecipitates, only the corresponding protein was detected. These results demonstrate the validity of the assay and indicate a specific interaction between MRP-L18 and 5S rRNA in vivo.
Figure 5.
Interaction of MRP-L18 with 5S rRNA in vivo analyzed by CLIP. (A) RNA isolated from the immunoprecipitates by either anti-MRP-L18 or aldolase antibodies (as indicated below the panels) obtained from HepG2 cells treated with formaldehyde. Two percent of input, wash, and protein A Sepharose flow-through fractions were loaded in the corresponding slots. RNA was separated in a 10% urea gel and analyzed by Northern hybridization with the 32P-labeled probes as indicated at the left (the list of the probes is in the Supplemental Material). (B) Western analysis of the immunoprecipitates. Two percent of the input and 5% of the immunoprecipitates were analyzed. (C) Analysis of the immunoprecipitates for the presence of various rRNAs by dot hybridization. The probes are indicated above the autoradiographs (and are listed in the Supplemental Material). The schema of the analyzed RNA dotted on the membrane is below the panel and its description is at the right. (w/o FA) Without formaldehyde treatment. The top row is a quantitative control with pure individual RNAs (T7-transcripts). The second row shows 20% of inputs (RNA isolated from an aliquot of lysed cells). The third row shows 20% of the protein A Sepharose washes. The fourth row shows the 50% of the RNA isolated from the immunoprecipitates.
We next attempted to check whether the same assay could provide further information concerning the partners of MRP-L18 and 5S rRNA. The precursor preMRP-L18 is expected to be present in low amounts in the cytosol, being rapidly imported and converted into the mature form in mitochondria, where it is presumably localized in mitochondrial ribosome large particles. We therefore analyzed the obtained immunoprecipitates for the presence of mitochondrial 12S and 16S rRNA by dot hybridization. In this experiment, immunoprecipitates were obtained by formaldehyde treatment of isolated mitochondria, in the hope of enriching mitochondria-related material. As one can see in Figure 5C, 5S rRNA was identified by this method as well, along with detectable amounts of both 12S and 16S rRNAs. As in the experiment above, 5.8S rRNA was not detected in any immunoprecipitate, nor were mitochondrial rRNAs detected in anti-aldolase ones, demonstrating the specificity of the assay. Minor amounts of 16S rRNA could be detected in the immunoprecipitates without cross-linking, which may be explained by the coprecipitation of large ribosomal subunits by anti-MRP-L18 antibodies. These results indicate a close association of all the three rRNA species, and therefore suggest that the imported 5S rRNA might be associated with the mitochondrial ribosome. The results obtained lead us to the more detailed investigation of the possible 5S rRNA association with mitochondrial ribosomes.
5S rRNA imported into mitochondria is associated with mitochondrial ribosomes
The question of possible integration of the imported 5S rRNA into mitochondrial mammalian ribosomes was debated from the very discovery of the import process (Magalhaes et al. 1998; Koc et al. 2001b). Although it seems generally accepted that the cytosolic 5S rRNA is absent from mammalian mitoribosomes, such a conclusion is based on only two pieces of experimental evidence (Koc et al. 2001b; Sharma et al. 2003). In the first study (Koc et al. 2001b), 39S large mitoribosome subunits obtained in low-magnesium conditions were used. It is known though, that, after such a treatment, 5S rRNA–protein complexes are quantitatively liberated from ribosomes of eukaryotes (Blobel 1971), bacteria (Horne and Erdmann 1972), and archaea (McDougall and Wittmann-Liebold 1994). It should be also noted that, although MRP-L18 was first identified in this study, it seems utterly depleted since only one peptide was found. (For comparison, a much milder and more rapid protocol of coimmunoprecipitation of mitochondrial ribosomes from human mitochondrial lysates allowed us to obtain preparations in which MRP-L18 is reproducibly identified as a major protein by at least three peptides [Rorbach et al. 2008; Richter et al. 2010].) The second study concerns cryo-electronic images of mammalian mitoribosomes obtained in a long isolation procedure followed by sorting of projections (Sharma et al. 2003). The resulting picture shows an abnormal morphology of the central protuberance, which, in particular, did not allow us to fit MRP-L18 and some other proteins into the electronic density map (Sharma et al. 2009).
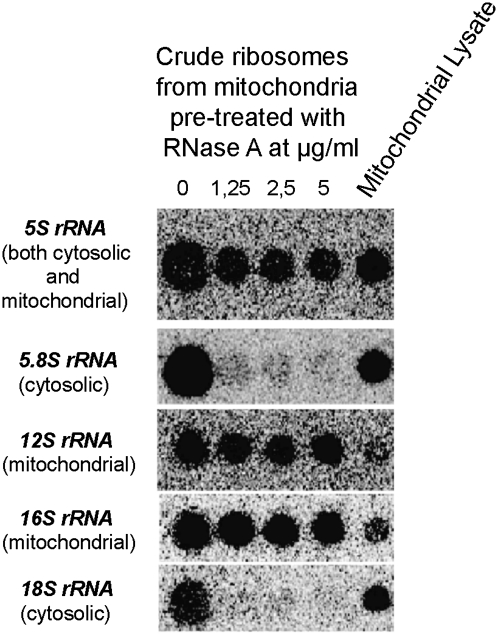
Our finding of the specific 5S rRNA-binding activity of MRP-L18 and the previously reported stoichiometry between the imported 5S rRNA and mitochondrial ribosomes in human cells (Entelis et al. 2001) encouraged us to readdress this question. To this end, we first used the standard protocol of isolation of mitoribosomes from rat liver. To get rid of the standard contaminant of mitochondrial preparations—cytosolic ribosomes attached to the outer membrane—we pretreated mitochondria with increasing concentrations of RNase A and then performed ribosome sedimentation from the cleared mitochondrial lysate. rRNA content analysis of the resulting preparations was then performed (Fig. 6), showing that, although 5.8S rRNA (large cytosolic subunit) and 18S rRNA (small cytosolic subunit) were quantitatively removed by RNase treatment, 5S rRNA as well as 12S and 16S mitochondrial rRNAs were perfectly preserved and cosedimented in a fraction corresponding to mitoribosomes (Koc et al. 2001b).
Figure 6.
Cytosolic 5S rRNA is detected in rat mitochondrial ribosomes. Dot blot Northern hybridization of RNAs isolated from crude rat liver mitoribosomal preparations, pretreated with various concentrations of RNase A. Hybridization probes are indicated at the left of each panel (see the Supplemental Material for sequences).
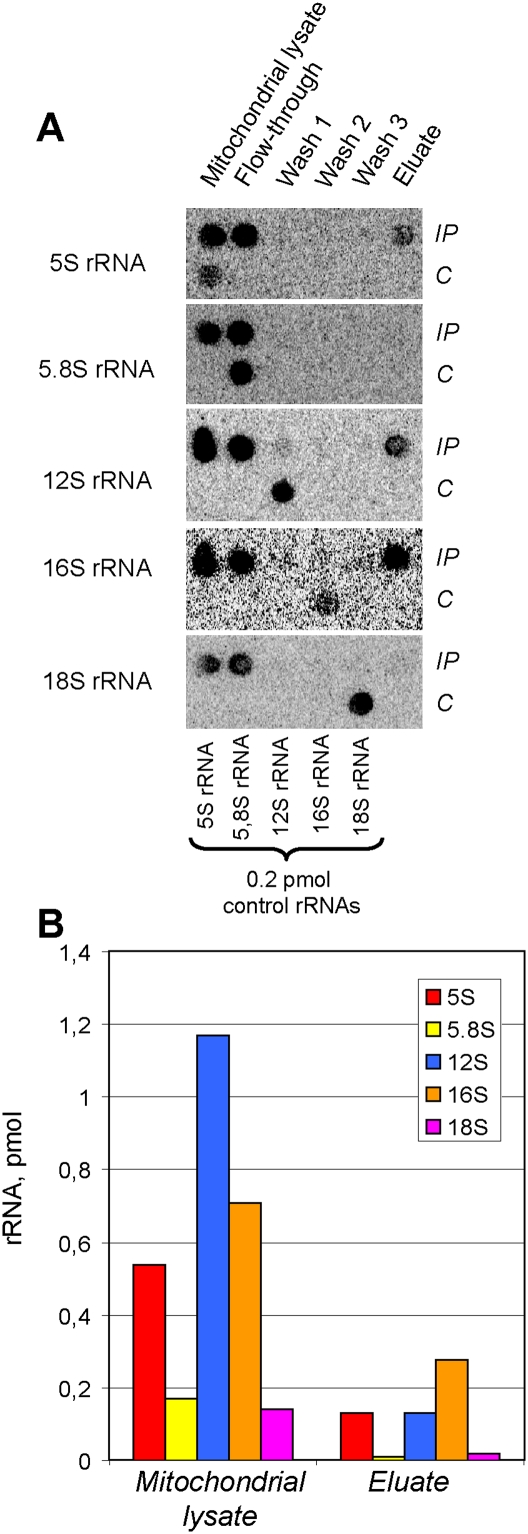
In a search for a milder isolation procedure, we turned to the coimmunoprecipitation protocol that appears to be optimal for detailed component analysis of functional mammalian mitoribosomes (Rorbach et al. 2008; Richter et al. 2010). We therefore isolated mitochondria from HEK293T cells expressing the Flag-tagged peptidyl hydrolase ICT1, a constitutive component of the mitochondrial large ribosomal subunit (Richter et al. 2010). The coimmunoprecipitate, containing at least several large subunit ribosomal proteins detected by mass spectrometry (Supplemental Table S1), was then analyzed for its rRNA content (Fig. 7). Once again, 5S, together with 12S and 16S rRNAs, was identified, while 5.8S and 18S, although present in the mitochondrial lysate, were not coimmunoprecipitated with mitochondrial ribosomes. The control experiment performed with the same cell line without induction of expression of Flag-ICT1 did not permit us to detect any 5S rRNA (or any other one tested) in the immunoprecipitate, indicating that the assay was protein-specific (Supplemental Fig. S4). Quantitative analysis indicates that the 16S rRNA was more abundant, which is consistent with the predominance of large mitoribosomal subunits in this preparation. 5S rRNA and 12S rRNA were present in the eluate in equimolar amounts (Fig. 7B), indicating that 5S rRNA can be dissociated from the large mitoribosomal subunit during purification and immunoprecipitation steps, but is detected in whole mitoribosomes in amounts comparable with 12S rRNA.
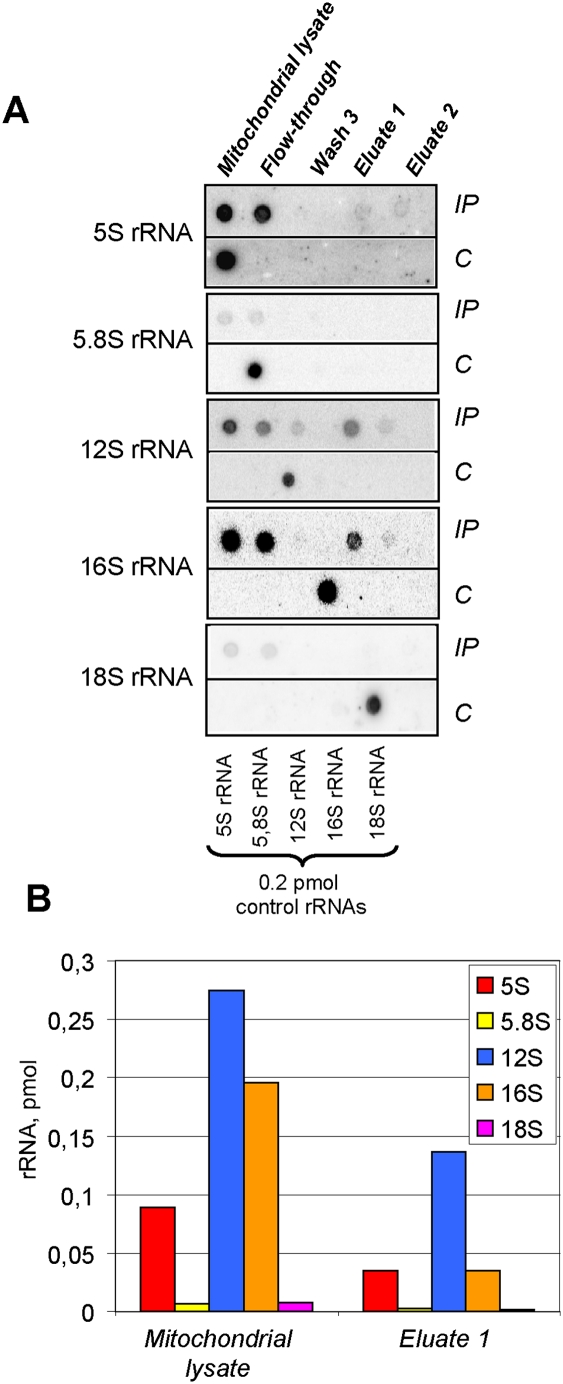
Figure 7.
Human mitoribosomes coimmunoprecipitated with ITC1-Flag contain cytosolic 5S rRNA. (A) Dot blot Northern hybridization of RNAs isolated from ICT1-Flag coimmunoprecipitated human mitochondrial ribosomes (IP, shown in the top lines). (C) rRNA transcripts used as quantitative controls (0.2 pmol) are dotted in the bottom line of each panel. Hybridization probes are indicated at the left. (B) Quantitative analysis of RNA contents in ICT1-Flag coimmunoprecipitated human mitochondrial ribosomes.
In order to validate the above result by an independent experiment, we analyzed ribosomes immunoprecipitated from HEK293T cells overexpressing another mitochondrial Flag-tagged ribosomal protein, MRP-S27, a part of a small ribosomal subunit (Koc et al. 2001a). The coimmunoprecipitated material, devoid of cytosolic contamination (5.8S and 18S rRNAs), contained all three mitochondrial rRNAs: 12S, 16S, and 5S (Fig. 8). In contrast to the ICT1-Flag immunoprecipitate, the 12S rRNA was largely more abundant, which is consistent with the increased presence of small ribosomal subunits in this preparation. Nevertheless, the amounts of 5S and 16S rRNAs in the eluate were equimolar. This result is in agreement with the analysis of the Flag-tagged ICT1 coimmunoprecipitate and favors the idea that cytosolic 5S rRNA can be present in mitochondrial ribosomes.
Figure 8.
Human mitoribosomes coimmunoprecipitated with MRP-S27-Flag contain cytosolic 5S rRNA. (A) Dot blot Northern hybridization of MRP-S27-Flag coimmunoprecipitates with rRNA-specific probes, as in Figure 6. (B) Quantification of the dot hybridization data.
We note that, according to the above data, 5S rRNA may be stably associated only with whole mitoribosomes, as confirmed by a strict equimolarity between immunoprecipitated rRNAs observed in both experiments. Thus, dissociation in subunits will probably result in a loss of this component. In fact, linear sucrose gradient sedimentation of mitoribosomes coimmunoprecipitated with MRP-S27 allowed us to detect 5S rRNA only in the peak corresponding most probably to whole monomitoribosomes (Supplemental Fig. S5). We can hypothesize that the interaction of MRP-L18 with proteins of the small subunit of the mitoribosome (shown for corresponding proteins in prokaryotic and eukaryotic ribosomes) (Ban et al. 2000; Ben-Shem et al. 2010) is needed to stabilize the MRP-L18–5S rRNA complex as a compound of the human mitochondrial ribosome.
Discussion
5S rRNA targeting to mammalian mitochondria: role of protein factors
One can hardly imagine the existence of an RNA molecule devoid of any protein partner in cells. Indeed, any RNA fate and function depend on interacting proteins that are essential for their stabilization, proper intracellular localization, and, in most cases, function. Although this paradigm has already become somewhat trivial, more and more examples of protein-dependent RNA biogenesis pathways do not cease to surprise us by their diversity and versatility. This is especially the case for mechanisms of RNA import into mitochondria described in a variety of eukaryotes (for review, see Tarassov et al. 2007; Salinas et al. 2008). Having emerged relatively late and independently in different taxa, these mechanisms are distinguished by the particularly inventive approaches that the cells use to move functional cytosolic RNA into the mitochondrial compartment. Import of 5S rRNA into mammalian mitochondria clearly stands apart in these studies, most of which concern tRNA relocalization events, beyond all doubt the most common case of RNA import (Duchene et al. 2009). This is why deciphering its mechanism and functional significance may become an important contribution in the whole theory of RNA localization in different compartments of the eukaryotic cell.
After the synthesis of eukaryotic 5S rRNA, its every relocalization event falls under the control of numerous protein partners (Fig. 9). Interestingly, the most important of them have pronounced RNA chaperone activities. Thus, 5S rRNA intracellular transport and even functioning are accompanied by changes of its conformation. For instance, newly synthesized 5S rRNA molecules are eventually bound by La and Ro proteins, which, thanks to their putative RNA chaperone activities, participate in “quality control” of transcripts (O'Brien and Wolin 1994; Shi et al. 1996). Once exported from the nucleus in complex with TFIIIA (Rudt and Pieler 1996), 5S rRNA changes partners again to ribosomal protein L5, which binds the molecule (Steitz et al. 1988). Upon the interaction, conformations of both partners suffer profound reorganization, leading to exposition of L5 nuclear localization signals and reimport into nucleus, then into the nucleolus (DiNitto and Huber 2003). Incorporation into nascent large ribosomal particles and even reassociation of ribosome subunits do not allow 5S rRNA to stay unchanged, since further conformational changes during these steps were reported (Shpanchenko et al. 1998). More recently, it was demonstrated that the very capacity of 5S rRNA to change its global structure is essential for performing its functions inside translating ribosomes (Kouvela et al. 2007). Thus, the chaperone events appear to reign over 5S rRNA in every aspect of its molecular fate.
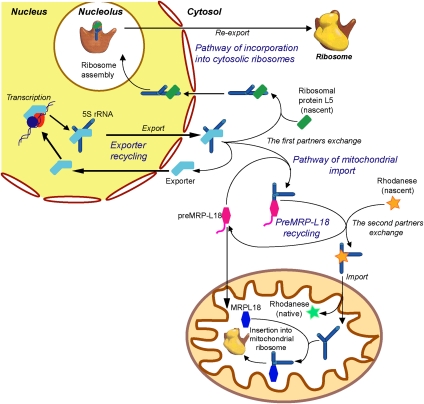
Figure 9.
Hypothetic mechanism of intracellular 5S rRNA traffic.
The scheme of intracellular traffic of mammalian 5S rRNA appeared even more complicated after its import into mitochondria had been discovered (Yoshionari et al. 1994; Magalhaes et al. 1998). The existence of two mutually exclusive destinations for the same RNA raised the question about the mechanism of its intracellular distribution. It appeared that, once again, a sequential chaperone-based vehicle was developed to remove a part of the 5S rRNA cytosolic pool from its usual “channeled” circuit and redirect it to the mitochondrial compartment (Fig. 9). Previously, we mapped the signals of mitochondrial localization of 5S rRNA (Fig. 1A), hypothesizing that they may match binding sites for protein factors involved in this pathway (Smirnov et al. 2008a). Indeed, recently, one of these factors was identified as the mitochondrial enzyme rhodanese (Smirnov et al. 2010). Having a strong and specific affinity to 5S rRNA, rhodanese was shown to bind it cotranslationally, just like ribosomal protein eL5 does (Lin et al. 2001). Functionally mimicking protein cochaperones involved in targeting of mitochondrial protein precursors to the outer membrane receptors, 5S rRNA thus ensures its own targeting to mitochondria. Recent discovery of another protein factor, obviously implicated at the stage of translocation through the inner mitochondrial membrane and identified as polynucleotide phosphorylase, substantially enriched our knowledge about protein factors needed for RNA import in human cells (Wang et al. 2010). However, the exact mechanism of RNA translocation through the mitochondrial membranes remained mostly unclear and awaited further elucidation.
It was shown that, for productive binding to rhodanese, 5S rRNA needs to be refolded into a “branched” conformation, suggesting the requirement of an upstream protein factor with pronounced chaperone activity (Smirnov et al. 2010). In the present study, this factor was identified as the precursor of mitochondrial ribosomal protein L18 (preMRP-L18). It appeared that this protein is ideally adapted for performing such a function. Having a strong and specific affinity to cytosolic 5S rRNA (Table 1), with a dissociation constant close to that usually observed for the eL5/5S rRNA interaction (Scripture and Huber 1995), preMRP-L18 can efficiently compete with ribosomal protein eL5 in order to intercept 5S rRNA molecules from the major circuit and redirect them to mitochondria (Fig. 9, step indicated as “the first partner exchange”).
Upon formation of the noncanonical complex with preMRP-L18, refolded 5S rRNA is rapidly discharged to the second import factor, rhodanese, both proteins thus functioning in a fast-rotating and unidirectional conveyor (Figs. 4, 9). Thus, the minimum 5S rRNA mitochondrial targeting protein vehicle has been established in this study, explaining how this cytosolic RNA can be redirected to mitochondria.
MRP-L18 and possible function of 5S rRNA in mammalian mitochondria
Since no 5S rRNA genes can be found in mammalian mitochondrial genomes, localization of cytosolic 5S rRNA inside the organelles provoked a debate about its possible involvement in mitochondrial translation. Until now, no attempts to find a 5S rRNA inside mammalian mitoribosomes were successful (Koc et al. 2001b; Sharma et al. 2003). Still, considerable evidence on its implication in mitochondrial protein synthesis has been accumulated. In particular, it was found that the number of 5S rRNA copies inside mitochondria matches that of mitochondrial ribosomes (Entelis et al. 2001). In our previous work, we have shown that knockdown of one of the 5S rRNA import factors, rhodanese, leads to not only a decrease of mitochondrial 5S rRNA level, but also the general fall of mitochondrial translation (Smirnov et al. 2010). Moreover, the most conservative 5S rRNA-binding partner, MRP-L18, was found to be a constituent of mammalian mitochondrial ribosomes (Koc et al. 2001b; Rorbach et al. 2008; Richter et al. 2010). It should be noted that its presence is well correlated with that of 5S rRNA inside the ribosome in all known taxa (Smits et al. 2007). It appears that, in a majority (although not all) of unicellular eukaryotes (and possibly all fungi), mitochondrial ribosomes are devoid of 5S rRNA. This component was successfully replaced by newly acquired mitochondria- and taxon-specific ribosomal proteins in Leishmania (see Sharma et al. 2009). No homologs of L18 were identified in yeast S. cerevisiae, Neurospora crassa, and protozoan mitochondria (Gan et al. 2002, 2006; Zikova et al. 2008; Sharma et al. 2009), confirming the correlation of L18’s presence inside the ribosome with that of 5S rRNA.
Here we show that human MRP-L18, being a prokaryotic-type 5S rRNA-binding protein, not only preserves its 5S rRNA-binding activity, but interacts efficiently with the cytosolic, eukaryotic-type 5S rRNA. From both structural and functional points of view, MRP-L18 represents quite a usual combinatory way of evolving ubiquitously found in the L18/eL5 family. Its core structure, composed of a β-sheet and a bundle of helices, appears to be well conserved and represents an ancient general RNA-binding domain (Yaguchi et al. 1984; Woestenenk et al. 2002). On the contrary, N-terminal extensions of L18/eL5 family proteins show a much greater diversity, making every member of the group unique and particularly well adapted for binding its proper substrate (Nazar et al. 1979; Newberry and Garrett 1980; Woestenenk et al. 2002). Always of moderate size, N-terminal extensions harbor elements responsible for specificity of interaction with 5S rRNA (note an especially dense interaction map for the N-terminal portion of the L18 protein in Fig. 1C), as well as some other functional elements like nuclear or mitochondrial localization signals or unstructured regions for chaperone-like binding to 5S rRNA (Fig. 1C; Supplemental Fig. S3; Rudt and Pieler 1996; Lin et al. 2001; DiNitto and Huber 2003). MRP-L18 has acquired a very specific N-terminal extension, which seems to be essential for the formation of a noncanonical complex with cytosolic 5S rRNA (Table 1).
This fact encouraged us to readdress the question of the 5S rRNA's presence inside mammalian mitoribosomes. Using milder and more rapid protocols of purification, we were able to show that it was indeed present in the rat liver and human mitochondrial ribosomes as a quasi-stoichiometric component, suggesting that previous unsuccessful attempts to localize it inside were compromised by a very labile association of this molecule with the core of the large subunit.
Thus, it appears that the entire 5S rRNA mitochondrial import pathway serves finally the same goal as its cytosolic counterpart: to direct a 5S rRNA molecule to the place of assembly of ribosomal subunits. Remarkable is the mirroring parallelism of both pathways, which depend on binding to either a eukaryotic or bacterial-type member of the same family of ribosomal proteins, inducing certain conformational changes in 5S rRNA and using their nucleolar or mitochondrial localization signals to deliver 5S rRNA to nascent ribosomes of the corresponding cellular compartment (Fig. 9). This represents the new example of an adaptation mechanism developed by eukaryotic cells to reinforce control over the mitochondrial genetic system. If one takes into account that the MRP-L18 protein is encoded in the genomes of most animals, including Caenorhabditis elegans (Koc et al. 2001b), it seems probable that this situation might be generalized over all of the kingdom.
What is the function of 5S rRNA inside mitoribosomes? Although a thorough analysis is needed to establish it directly, one may suppose that it does not differ much from the canonical one (Bogdanov et al. 1995; Smith et al. 2001; Kiparisov et al. 2005). Indeed, although the large mitochondrial 16S rRNA suffered a drastic reduction in size and an extensive loss of structural elements compared with its prokaryotic counterpart, every single helix implicated in the 5S rRNA-dependent intraribosomal signal transduction (H80, H89, H39, H91, H92, and H95) is perfectly preserved, suggesting that this regulatory system still exists in the mitochondrial large ribosomal subunit.
Materials and methods
5S rRNA manipulations
All of the 5S rRNA variants used are described in detail in Smirnov et al. (2008a) and are also listed in Table 1. Corresponding PCR-generated genes were transcribed in vitro using the T7 RiboMAX Express Large-Scale RNA Production System (Promega), resulting in transcripts purified by denaturing urea-PAGE. For the branched and standard conformations, 5S rRNA was incubated for 1 min at 100°C in 10 μL of either 1 mM MgCl2 (to obtain “compact” 5S rRNAMg) or 1 mM EDTA (to obtain “branched” 5S rRNAEDTA) (Smirnov et al. 2010). Then, the RNA was cooled to 4°C and immediately loaded on the running native 10% PAAG (0.5× TBE, 5% glycerol) to minimize rapid conformational changes of the RNA.
In all experiments in which it was not specified, the 5S rRNAMg form was used.
Rhodanese manipulations
Bovine liver rhodanese (Sigma) was stored in 50 mM Tris-HCl, 20 mM DTT, and 50 mM sodium thiosulfate (pH 7) at −80°C. Urea-denatured rhodanese was prepared and the aggregation assay was performed as described in Silberg et al. (1998) and Smirnov et al. (2010).
Precursor and mature forms of MRP-L18
Precursor and mature forms of MRP-L18 as well as its N-terminal (residues 1–79) and C-terminal (residues 80–180) portions were His6-tagged on their C termini, expressed, and purified from E. coli cells.
Import of 5S rRNA into isolated human mitochondria
Human mitochondria were isolated from HepG2 cells as described in Gaines and Attardi (1984) and Entelis et al. (2001) using buffer containing 0.6 M sorbitol, 10 mM HEPES-NaOH (pH 7.0), and 1 mM EDTA. The standard RNA import assay was performed as described (Entelis et al. 2001). For this, mitochondria were incubated with [32P]-γ-ATP-labeled T7 transcripts in the presence of purified rhodanese and/or (pre)MRP-L18, ATP, and a phosphoenolpyruvate kinase system for 10 min at 37°C (linear phase of RNA uptake). The RNA uptake step was followed by mitochondria repurification, RNases treatment to eliminate all of the external RNA, washing, and RNA isolation from repurified mitochondria. Imported RNAs were analyzed by PAGE in 10% polyacrylamide gels containing 8 M urea; quantification of results was done by a Typhoon-Trio scanner using ImageQuantTools software (GE Healthcare). To estimate the import efficiency, 50 fmol of corresponding 32P-labeled RNA was loaded on the same gel. All manipulations with mitochondria, up to the RNA isolation step, normally take at least 1 h, so the amount of imported RNA detected by gel scanning should correspond to labeled RNA that had been penetrated into the mitochondrial matrix and did not degrade there in 1 h. In all experiments, negative controls without mitochondria or proteins were performed to confirm the absence of protein–RNA aggregation.
EMSA
Purified [32P]-labeled RNA (0.1–10 nM) was denatured at 100°C in the presence of 1 mM MgCl2, then slowly cooled to room temperature. It was then incubated with rhodanese and/or (pre)MRP-L18 in 20 mM Tris-HCl, 100 mM NaCl, and 5 mM MgCl2 (pH 7.0) for 10–15 min. The resulting complexes were analyzed by electrophoresis in 8% PAAG, 0.5× TBE, and 5% glycerol in 0.5× TBE buffer at 10 V/cm at 4°C followed by Typhoon-Trio (GE Healthcare) scanning and quantification.
The standard Scatchard plot construction and analysis are described in Henis and Levitzki (1976). For this, several (at least six) standard EMSA reactions were performed in which the protein concentration was constant (0.45 μM) and the concentration of labeled RNA varied in a range from 10 nM to 12.5 pM. Resulting autoradiographs were analyzed by an ImageQuantTL package, and the signals corresponding to bound and free RNA were measured. The Scatchard plot is a linearized form of binding curve described by the equation
where [Bound RNA] and [Free RNA] are molar equilibrium concentrations of the RNA–protein complex and free RNA, respectively, and a is a constant. Thus, the linear regression of experimental data allows a calculation of the dissociation constant Kd.
In some cases, Scatchard plots were nonlinear: A concave curve indicates the presence in protein preparation of multiple binding sites with different affinities; in this case, the accurate determination of Kd is difficult, and curve decomposition usually allows only approximate Kd estimation. Here we provide only average estimated values for such cases. An asymptotic approach of the curve to the X-axis at high [Bound RNA] values is a natural trait of Scatchard plots caused by the impossibility of accurately measuring the ordinate in this area because of too low [Free RNA]; this area was excluded from analysis. At very low [Bound RNA] values, the ordinate tends to fluctuate; this is again a natural consequence of the impossibility of accurately measuring [Bound RNA], and this area was also excluded.
The specially designed EMSA assay for MRP-L18 chaperone capacity analysis was set up as follows: Mg-folded 32P-labeled 5S rRNA (7.5 nM) was incubated with recombinant preMRP-L18 (0.6 μM) for 20 min at 20°C, and the 50-fold excess of unlabeled Mg-5S rRNA was added. The chase continued for 1–5 min at 20°C, and aliquots were loaded on the running native 10% PAAG (0.5× TBE, 5% glycerol) to minimize rapid conformational changes of the RNA.
Silencing of the MRPL-L18 gene
Silencing of the MRPL-L18 gene was ensured by consecutive transfections of HepG2 cells spaced by 3 d with a mixture of three cognate siRNAs (Santa Cruz Biotechnology) in accordance with the manufacturer's protocol. For control, cells were transfected with siRNAs against Luciferase mRNA, as described in Kolesnikova et al. (2004), or mock-transfected (without siRNA). The level of preMRP-L18 expression was tested by Western blot analysis with MRP-L18-directed polyclonal mouse antibodies (Abcam). In agreement with the manufacturer's specifications, these antibodies recognize two proteins in total cellular lysate, with the upper band being nonspecific and the lower one corresponding to MRP-L18. Therefore, only the lower one was quantified in silencing experiments (Fig. 3C). Mitochondria were isolated and purified as described in Gaines and Attardi (1984), and treated with RNase A and digitonin to remove nonspecifically attached RNAs. Total and mitochondrial RNAs were isolated with TRIZol reagent (Invitrogen) and analyzed by Northern blot hybridization (see the Supplemental Material for hybridization probes).
Predictions of the tertiary structure of MRP-L18
Predictions of the tertiary structure of MRP-L18 were performed with the help of Geno3D2 (Combet et al. 2002), 3D-JIGSAW (Contreras-Moreira and Bates 2002), Psipred (Bryson et al. 2005), and Porter (Pollastri and McLysaght 2005) software.
CLIP
CLIP was performed essentially as described in Mei et al. (2010) with either whole cultured cells or isolated mitochondria. HepG2 human cells cultured in DMEM with 10% of FBS at standard conditions were harvested and PBS-washed. Low-concentration (0.2%) formaldehyde treatment was performed for 10 min at 20°C in order to minimize nonspecific cross-linking (Niranjanakumari et al. 2002). The cross-linking was quenched in 150 mM glycine (pH 7.5). Extracts were prepared in immunoprecipitation buffer (20 mM Tris at pH 7.8, 500 mM NaCl, 2.5 mM MgCl2) containing 1% of the Empigen BB detergent (Choi and Dreyfuss 1984). Immunoprecipitation was performed after 2 h of incubation with the appropriate antibody followed by protein A Sepharose treatment. To further analyze RNAs, immunoprecipitates were heated for 30–45 min at 75°C, and RNA was extracted by Trizol reagent and further submitted to Northern hybridization with appropriate 32P-labeled oligonucleotide probes. To analyze proteins, aliquots of the immunoprecipitates were analyzed by standard SDS-PAGE followed by Western analysis. Similar experiments were also performed with isolated mitochondria.
Isolation and component analysis of mammalian mitochondrial ribosomes
Isolation and component analysis of mammalian mitochondrial ribosomes were performed as described in Spremulli (2007) and Richter et al. (2010). Briefly, mitochondria were isolated from livers of freshly sacrificed 4-wk-old rats as described in Entelis et al. (2001) using Mg2+-free buffer containing 0.6 M sorbitol, 10 mM HEPES-NaOH (pH 7.0), and 1 mM EDTA; treated with RNase A; repurified; and lysed with 1.6% Triton X-100 in 0.26 mM sucrose, 40 mM KCl, 15 mM MgCl2, 14 mM Tris-HCl (pH 7.5), 0.8 mM EDTA, 50 μM spermine, 50 μM spermidine, 5 mM β-mercptoethanol, 0.25% PMSF, and 50 U/mL RNaseOUT (Invitrogen). The cleared lysate was layered on a 1 M sucrose cushion in 100 mM KCl, 15 mM MgCl2, 20 mM triethanolamine (pH 7.5), 5 mM β-mercptoethanol, 1% Triton X-100, 0.25 mM PMSF, and 40 U/mL RNaseOUT and centrifuged at 100,000g for 7 h. The resulting pellets were washed briefly with the same buffer, and RNAs were isolated with TRIzol reagent (Invitrogen).
For coimmunoprecipitation studies, HEK293T cell lines stably transfected with either ICT1-Flag or MRP-S27-Flag gene were used. Mitochondria were isolated from induced cells using Mg2+-free buffer, treated with proteinase K and digitonin as described in Rorbach et al. (2008), and finally lysed with 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 20 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, proteinase inhibitor cocktail (Roche), and 50 U/mL RNaseOUT. The cleared lysate was incubated with anti-Flag M2-agarose affinity gel, and the immunoprecipitation, washing, and elution steps were done according to the manufacturer's protocol (Sigma Aldrich). Immunoprecipitated material was analyzed by sucrose gradient centrifugation as described elsewhere (Richter et al. 2010). RNAs isolated from material eluted from an anti-Flag affinity gel were analyzed by dot blot Northern hybridization using short T7 transcripts corresponding to cytosolic and mitochondrial rRNA sequences as quantitative controls.
MALDI-MS and LC-MS/MS analyses
MALDI-MS and LC-MS/MS analyses were performed on a BIFLEX III MALDI-TOF (Bruker Daltonics) by standard procedures.
North–Western blot hybridization
North–Western blot hybridization with [32P]-labeled human 5S rRNA was carried out in the Import buffer, as in Entelis et al. (2006). HepG2 mitochondrial proteins (20 μg) were separated on 10% SDS-PAAG and blotted to a nitrocellulose membrane. The membrane was incubated in 0.1 M Tris-HCl, 20 mM KCl, 2.5 mM MgCl2, and 0.1% Nonidet P-40 (pH 7.5) for 1 h at 4°C with stirring, then washed several times with the same solution and blocked in 10 mM Tris-HCl (pH 7.5), 5 mM Mg(CH3COO)2, 2 mM dithiothreitol, 5% BSA, and 0.01% Triton X-100 for 5 min at 4°C. Then, the membrane was incubated for 2 h at 37°C in the Import buffer containing 1 nM labeled human 5S rRNA, washed with the same buffer without 5S rRNA, and autoradiography-analyzed by Typhoon-Trio (GE Healthcare) scanning and quantification.
Acknowledgments
We thank R.N. Lightowlers, R. Richter, and J. Rorbach (Newcastle University, UK) for providing the ICT1-Flag and MRPS27-Flag constructs; P. Hammann (IBMC-CNRS, Strasbourg, France) for mass spectrometry analysis; P. Romby and S. Marzi (IBMC-CNRS, Strasbourg, France) for help in setting up ribosome purification; and A.-M. Heckel-Mager (UMR 7156, CNRS-Université de Strasbourg, France) for extensive technical assistance. This work was supported by the CNRS, Université de Strasbourg, Moscow State University, AFM (Association Française contre les Myopathies), ANR (Agence Nationale de la Recherche), FRM (Fondation pour la Recherche Médicale), and ARCUS (cooperation program Alsace-Russia-Ukraine).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.624711.
Supplemental material is available for this article.
References
- Aoyama K, Tanaka T, Hidaka S, Ishikawa K 1984. Binding sites of rat liver 5S RNA to ribosomal protein L5. J Biochem 95: 1179–1186 [DOI] [PubMed] [Google Scholar]
- Attardi G, Ojala D 1971. Mitochondrial ribosome in HeLa cells. Nat New Biol 229: 133–136 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Blobel G 1971. Isolation of a 5S RNA–protein complex from mammalian ribosomes. Proc Natl Acad Sci 68: 1881–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN 1995. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol 73: 869–876 [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT 2005. Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36–W38 doi: 10.1093/nar/gki410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaulk S, Lu J, Tan K, Arthur DC, Edwards RA, Frost LS, Joachimiak A, Glover JN 2011. N. meningitidis 1681 is a member of the FinO family of RNA chaperones. RNA Biol 7: 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G 1984. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol 99: 1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Jambon M, Deleage G, Geourjon C 2002. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics 18: 213–214 [DOI] [PubMed] [Google Scholar]
- Contreras-Moreira B, Bates PA 2002. Domain fishing: a first step in protein comparative modelling. Bioinformatics 18: 1141–1142 [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Huber PW 2001. A role for aromatic amino acids in the binding of Xenopus ribosomal protein L5 to 5S rRNA. Biochemistry 40: 12645–12653 [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Huber PW 2003. Mutual induced fit binding of Xenopus ribosomal protein L5 to 5S rRNA. J Mol Biol 330: 979–992 [DOI] [PubMed] [Google Scholar]
- Duchene AM, Pujol C, Marechal-Drouard L 2009. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet 55: 1–18 [DOI] [PubMed] [Google Scholar]
- Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA 2001. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. J Biol Chem 276: 45642–45653 [DOI] [PubMed] [Google Scholar]
- Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I 2006. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev 20: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto H, Taguchi A, Itoh T, Morinaga T 2000. 5S rRNA binding proteins from the hyperthermophilic archaeon, Pyrococcus furiosus. FEBS Lett 486: 195–199 [DOI] [PubMed] [Google Scholar]
- Gaines G, Attardi G 1984. Highly efficient RNA-synthesizing system that uses isolated human mitochondria: new initiation events and in vivo-like processing patterns. Mol Cell Biol 4: 1605–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Kitakawa M, Yoshino K, Oshiro N, Yonezawa K, Isono K 2002. Tag-mediated isolation of yeast mitochondrial ribosome and mass spectrometric identification of its new components. Eur J Biochem 269: 5203–5214 [DOI] [PubMed] [Google Scholar]
- Gan X, Arita K, Isono S, Kitakawa M, Yoshino K, Yonezawa K, Kato A, Inoue H, Isono K 2006. Identification and comparative analysis of the large subunit mitochondrial ribosomal proteins of Neurospora crassa. FEMS Microbiol Lett 254: 157–164 [DOI] [PubMed] [Google Scholar]
- Gongadze GM, Perederina AA, Meshcheriakov VA, Fedorov RV, Moskalenko SE, Rak AV, Serganov AA, Shcherbakov DV, Nikonov SV, Garber MB 2001. The Thermus thermophilus 5S rRNA-protein complex: identifications of specific binding sites for proteins L5 and L18 in 5S rRNA. Mol Biol (Mosk) 35: 610–616 [PubMed] [Google Scholar]
- Henis YI, Levitzki A 1976. An analysis on the slope of Scatchard plots. Eur J Biochem 71: 529–532 [DOI] [PubMed] [Google Scholar]
- Horne JR, Erdmann VA 1972. Isolation and characterization of 5S RNA–protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol Gen Genet 119: 337–344 [DOI] [PubMed] [Google Scholar]
- Huber PW, Wool IG 1984. Nuclease protection analysis of ribonucleoprotein complexes: use of the cytotoxic ribonuclease α-sarcin to determine the binding sites for Escherichia coli ribosomal proteins L5, L18, and L25 on 5S rRNA. Proc Natl Acad Sci 81: 322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiparisov S, Petrov A, Meskauskas A, Sergiev PV, Dontsova OA, Dinman JD 2005. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Mol Genet Genomics 274: 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL 2001a. Identification of four proteins from the small subunit of the mammalian mitochondrial ribosome using a proteomics approach. Protein Sci 10: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL 2001b. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969 [DOI] [PubMed] [Google Scholar]
- Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I 2004. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet 13: 2519–2534 [DOI] [PubMed] [Google Scholar]
- Kouvela EC, Gerbanas GV, Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL 2007. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine. Nucleic Acids Res 35: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs D, Rakacs M, Agoston B, Lenkey K, Semrad K, Schroeder R, Tompa P 2009. Janus chaperones: assistance of both RNA- and protein-folding by ribosomal proteins. FEBS Lett 583: 88–92 [DOI] [PubMed] [Google Scholar]
- Lin E, Lin SW, Lin A 2001. The participation of 5S rRNA in the co-translational formation of a eukaryotic 5S ribonucleoprotein complex. Nucleic Acids Res 29: 2510–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes PJ, Andreu AL, Schon EA 1998. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol Biol Cell 9: 2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall J, Wittmann-Liebold B 1994. Comparative analysis of the protein components from 5S rRNA.protein complexes of halophilic archaebacteria. Eur J Biochem 221: 779–785 [DOI] [PubMed] [Google Scholar]
- Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X 2010. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 37: 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar RN, Willick GE, Matheson AT 1979. The 5 S RNA·protein complex from an extreme halophile, Halobacterium cutirubrum. Studies on the RNA–protein interaction. J Biol Chem 254: 1506–1512 [PubMed] [Google Scholar]
- Newberry V, Garrett RA 1980. The role of the basic N-terminal region of protein L18 in 5S RNA–23S RNA complex formation. Nucleic Acids Res 8: 4131–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA 2002. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods 26: 182–190 [DOI] [PubMed] [Google Scholar]
- O'Brien TW 2002. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286: 73–79 [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Wolin SL 1994. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev 8: 2891–2903 [DOI] [PubMed] [Google Scholar]
- Pace B, Stahl DA, Pace NR 1984. The catalytic element of a ribosomal RNA-processing complex. J Biol Chem 259: 11454–11458 [PubMed] [Google Scholar]
- Pollastri G, McLysaght A 2005. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21: 1719–1720 [DOI] [PubMed] [Google Scholar]
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM 2010. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J 29: 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA, et al. 2008. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 36: 5787–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudt F, Pieler T 1996. Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO J 15: 1383–1391 [PMC free article] [PubMed] [Google Scholar]
- Salinas T, Duchene AM, Marechal-Drouard L 2008. Recent advances in tRNA mitochondrial import. Trends Biochem Sci 33: 320–329 [DOI] [PubMed] [Google Scholar]
- Scripture JB, Huber PW 1995. Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5 S rRNA. The major determinants of recognition are located in helix III-loop C. J Biol Chem 270: 27358–27365 [DOI] [PubMed] [Google Scholar]
- Scripture JB, Huber PW 2011. Binding site for Xenopus ribosomal protein L5 and accompanying structural changes in 5S rRNA. Biochemistry 50: 3827–3839 [DOI] [PubMed] [Google Scholar]
- Semrad K, Green R, Schroeder R 2004. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA 10: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115: 97–108 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK 2009. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci 106: 9637–9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, O'Brien CA, Van Horn DJ, Wolin SL 1996. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA 2: 769–784 [PMC free article] [PubMed] [Google Scholar]
- Shpanchenko OV, Dontsova OA, Bogdanov AA, Nierhaus KH 1998. Structure of 5S rRNA within the Escherichia coli ribosome: iodine-induced cleavage patterns of phosphorothioate derivatives. RNA 4: 1154–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg JJ, Hoff KG, Vickery LE 1998. The Hsc66–Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK–DnaJ–grpE system. J Bacteriol 180: 6617–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Tarassov I, Mager-Heckel AM, Letzelter M, Martin RP, Krasheninnikov IA, Entelis N 2008a. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA 14: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov AV, Entelis NS, Krasheninnikov IA, Martin R, Tarassov IA 2008b. Specific features of 5S rRNA structure—its interactions with macromolecules and possible functions. Biochemistry (Mosc) 73: 1418–1437 [DOI] [PubMed] [Google Scholar]
- Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I 2010. Mitochondrial enzyme rhodanese is essential for 5S ribosomal RNA import into human mitochondria. J Biol Chem 285: 30792–30803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Meskauskas A, Wang P, Sergiev PV, Dinman JD 2001. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 21: 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ 2007. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35: 4686–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spremulli LL 2007. Large-scale isolation of mitochondrial ribosomes from mammalian tissues. Methods Mol Biol 372: 265–275 [DOI] [PubMed] [Google Scholar]
- Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T 1988. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol 106: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J 2003. 5 S rRNA: structure and interactions. Biochem J 371: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Kamenski P, Kolesnikova O, Karicheva O, Martin RP, Krasheninnikov IA, Entelis N 2007. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell Cycle 6: 2473–2477 [DOI] [PubMed] [Google Scholar]
- Tompa P, Csermely P 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175 [DOI] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, et al. 2010. PNPASE regulates RNA import into mitochondria. Cell 142: 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woestenenk EA, Gongadze GM, Shcherbakov DV, Rak AV, Garber MB, Hard T, Berglund H 2002. The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold. Biochem J 363: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M, Rollin CF, Roy C, Nazar RN 1984. The 5S RNA binding protein from yeast (Saccharomyces cerevisiae) ribosomes. An RNA binding sequence in the carboxyl-terminal region. Eur J Biochem 139: 451–457 [DOI] [PubMed] [Google Scholar]
- Yoshionari S, Koike T, Yokogawa T, Nishikawa K, Ueda T, Miura K, Watanabe K 1994. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS Lett 338: 137–142 [DOI] [PubMed] [Google Scholar]
- Zikova A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart K 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics 7: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]