Figure 1.
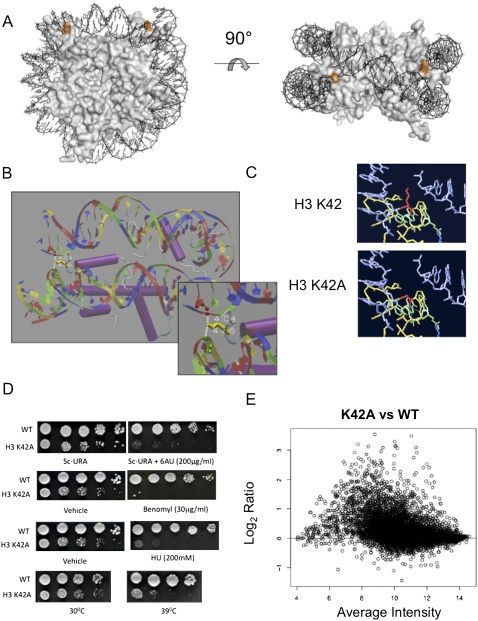
Histone H3-K42A is a pleiotropic mutation, which disrupts a structurally important nucleosome surface. (A) The crystal structure of the S. cerevisiae nucleosome highlighting histone H3 Lys 42 (orange) positioned at the DNA entry and exit points. The figure was generated using PYMOL. (B) Nucleosome crystal structure surrounding H3-K42 (yellow), indicating hydrogen bonds between the lysine side chain and DNA. (C) Predicted structures of H3-K42A mutation in its nucleosomal context. (Red) Residue 42; (blue) DNA; (green/yellow) histone H3. Shown is the single rotamer that exists for this mutation. Graphics were generated using Swiss PDB viewer. (D) Growth assays were undertaken for the indicated strains as described in Materials and Methods to detect defects in transcription elongation, cell cycle, and DNA repair, and to monitor temperature sensitivity at 39°C. (E) An MVA plot indicating gene expression changes versus spot intensity of normalized microarray data of K42A versus wild-type (WT) histone H3-expressing cells.