Abstract
Successful DNA replication and packaging of newly synthesized DNA into chromatin are essential to maintain genome integrity. Defects in the DNA template challenge genetic and epigenetic inheritance. Unfortunately, tracking DNA damage responses (DDRs), histone deposition, and chromatin maturation at replication forks is difficult in mammalian cells. Here we describe a technology called iPOND (isolation of proteins on nascent DNA) to analyze proteins at active and damaged replication forks at high resolution. Using this methodology, we define the timing of histone deposition and chromatin maturation. Class 1 histone deacetylases are enriched at replisomes and remove predeposition marks on histone H4. Chromatin maturation continues even when decoupled from replisome movement. Furthermore, fork stalling causes changes in the recruitment and phosphorylation of proteins at the damaged fork. Checkpoint kinases catalyze H2AX phosphorylation, which spreads from the stalled fork to include a large chromatin domain even prior to fork collapse and double-strand break formation. Finally, we demonstrate a switch in the DDR at persistently stalled forks that includes MRE11-dependent RAD51 assembly. These data reveal a dynamic recruitment of proteins and post-translational modifications at damaged forks and surrounding chromatin. Furthermore, our studies establish iPOND as a useful methodology to study DNA replication and chromatin maturation.
Keywords: DNA replication, chromatin, DNA damage response, H2AX, histone acetylation, EdU, click chemistry
In human cells, more than 6 billion base pairs of DNA need to be replicated and packaged into chromatin every cell division cycle. Failures lead to mutation, epigenetic changes, and other chromosomal aberrations that ultimately cause diseases such as cancer. DNA replication is coordinated with chromatin assembly (Probst et al. 2009). The replisome, containing the proteins necessary to complete replication, is a dynamic machine that must work with speed and precision. DNA lesions, insufficient nucleotides, and other types of replication stress cause fork stalling. In these circumstances, the DNA damage response (DDR) mobilizes repair activities to stabilize the fork, resolve the problem, and complete DNA synthesis (Harper and Elledge 2007; Cimprich and Cortez 2008).
The DDR to replication stress is poorly understood in comparison with the response to double-strand breaks (DSBs). For example, there are extensive modifications to the chromatin surrounding a DSB, including destabilization of nucleosomes, chromatin remodeling, and histone post-translational modifications (Morrison and Shen 2009; van Attikum and Gasser 2009; Rossetto et al. 2010; Venkitaraman 2010). These changes increase access to the repair machinery and recruit proteins involved in repair and DDR signaling. The extent to which chromatin changes at a stalled fork mimic those at a DSB is unknown.
Replication provides a unique landscape and set of challenges compared with a DSB. The immediate vicinity of the replisome lacks nucleosomes. Also, half of the histones on the nascent DNA are newly synthesized and require changes in post-translational modifications to restore the proper chromatin structure. Finally, several mechanisms exist to recover stalled replication forks, which necessitate the recruitment of multiple enzymatic activities and, perhaps, different chromatin changes.
The difference in our knowledge of the responses at stalled forks compared with DSBs is due primarily to the increased technical challenges of studying replication stress. For example, several investigators have used site-specific DSBs combined with chromatin immunoprecipitation (ChIP) to examine proteins at breaks with high resolution (Rudin and Haber 1988; Rodrigue et al. 2006; Soutoglou et al. 2007; Berkovich et al. 2008). Thus far, site-specific analysis of active and stalled replisomes in mammalian cells has not been achieved. We addressed this technical limitation by developing the iPOND (isolation of proteins on nascent DNA) methodology. iPOND permits the isolation and analysis of proteins at active, stalled, and collapsed replication forks. It can also probe the changes that accompany chromatin deposition and maturation following DNA synthesis. We demonstrate the power of iPOND by defining the dynamics of proteins and post-translational modifications in the replisome and on the newly deposited chromatin.
Results
Development of iPOND
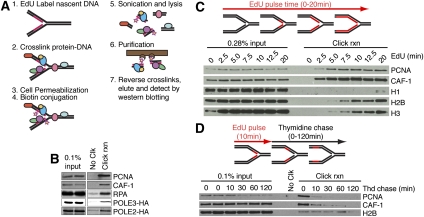
Tracking the location of any single replisome in a mammalian cell is not possible, limiting the utility of ChIP-based technologies. To overcome this technical limitation, we used the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) (Salic and Mitchison 2008), which contains an alkyne functional group. Covalent linkage to a biotin-azide using click chemistry (Moses and Moorhouse 2007) facilitates single-step purification of the EdU-labeled nascent DNA and associated proteins at replication forks (Fig. 1A).
Figure 1.
Development of the iPOND technology. (A) iPOND begins by adding EdU to cultured cells. The cells are then treated with formaldehyde to cross-link protein–DNA complexes, washed, and permeabilized with detergent. Copper catalyzes the cycloaddition of biotin-azide to the EdU-labeled DNA. The cells are then lysed in denaturing conditions with sonication. The biotin-labeled DNA–protein complexes are purified using streptavidin-coated beads, cross-links are reversed, and the eluted proteins are analyzed by immunoblotting or other methods like mass spectrometry. (B) Cells were incubated with EdU for 10 min prior to performing iPOND. Cells expressing POLE2-HA or POLE3-HA were used to detect these proteins with the HA antibody. (C) Cells were incubated in EdU-containing medium for increasing times prior to performing the iPOND protocol. (D) Cells were incubated with EdU for 10 min. The EdU-containing medium was removed and cells were washed once before incubating for increasing times in medium containing 10 μM thymidine prior to performing iPOND. In all experiments, the No Clk control is the input sample in the first lane processed with no biotin-azide.
To validate this methodology we first asked whether we could detect replisome proteins. We labeled cells with EdU for 10 min then performed iPOND. We detected proliferating cell nuclear antigen (PCNA), chromatin assembly factor 1 (CAF-1), replication protein A (RPA), and two subunits of polymerase ε (Fig. 1B). These results indicate that iPOND can purify replisome proteins, including those indirectly bound to DNA such as CAF-1 (Shibahara and Stillman 1999). Furthermore, they indicate that iPOND is a highly sensitive methodology. We are able to detect proteins such as POLE2 and POLE3, which are expected to be at a density of only one or two molecules per fork (Fig. 1B). Thus, unlike immunofluorescence, iPOND does not require high concentrations of proteins within a small nuclear region to track protein localization. Of note, proteins not present at replication forks, such as GAPDH, are not detectable in iPOND captures (data not shown).
In time-course experiments, we detected PCNA and CAF-1 after a 2.5-min pulse of EdU, histones H2B and H3 after 5 min, and the linker histone H1 at 20 min after EdU addition (Fig. 1C). Thus, with short labeling times, we selectively purify proteins at the replication fork, and longer labeling times permit analysis of chromatin assembly. The order of histone deposition supports previous fractionation data indicating that H1 is added 10–20 min after DNA replication to create higher-order chromatin structures (Worcel et al. 1978).
The resolution of this technique depends on the length of the EdU pulse, the rate of DNA synthesis, and the size of the DNA fragments generated after cell lysis. In practice, the first two parameters are the most important, since we consistently obtain DNA fragments of ∼150 base pairs (bp) (Supplemental Fig. 1). In mammalian cells, the rate of DNA synthesis varies between 0.75 and 2.5 kb/min (Herrick and Bensimon 2008). Thus, a 2.5-min EdU pulse labels ∼2–6 kb, although this is likely a significant overestimation, since EdU must enter the cell and be phosphorylated before incorporation into DNA. Thus, iPOND resolution is on the order of a few thousand base pairs.
Importantly, iPOND can be combined with pulse-chase methods to track how proteins assemble and disassemble from a nascent DNA segment with high spatial and temporal resolution. Increasing chase times monitor DNA-associated proteins at greater and greater distances from the moving fork. In these experiments, histone levels remain constant, indicating that the procedure effectively captures a maturing chromatin segment of constant length (Fig. 1D). However, PCNA and CAF-1 levels purified with the EdU-labeled segment decline with a half-life of considerably <10 min of chase time (Fig. 1D). These data indicate that iPOND isolates chromatin-associated proteins specifically located at the replication fork, and are consistent with rapid unloading of PCNA and CAF-1 once Okazaki fragment DNA synthesis is complete.
Analysis of chromatin maturation using iPOND
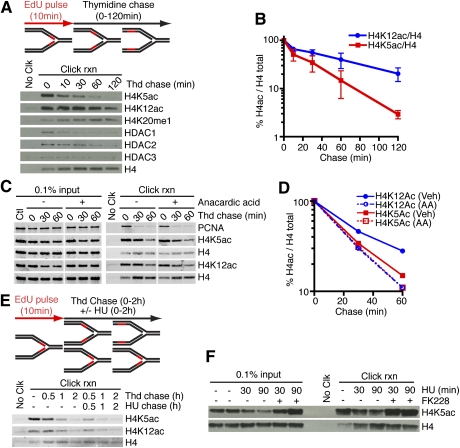
Maturation of the new chromatin requires addition and removal of histone post-translational modifications. Newly synthesized histone H4 is acetylated on two lysines (5 and 12), and these evolutionarily conserved marks are removed after deposition (Sobel et al. 1995; Taddei et al. 1999). Our time course experiments indicate that acetylated H4K5 (H4K5ac) is removed rapidly and H4K12ac deacetylation is slightly delayed (Fig. 2A,B). The delay in K12 deacetylation could be due to the activity of chromatin-associated histone acetyltransferases (HATs) that promote the acetylation of this site in some chromatin domains. Indeed, in the presence of the nonselective HAT inhibitor anacardic acid, the rate of H4K12 deacetylation becomes identical to H4K5, with a half-life of <20 min (Fig. 2C,D).
Figure 2.
HDACs are enriched at replication forks and deacetylate newly deposited histone H4 regardless of fork movement. (A–E). Cells were labeled with EdU for 10 min followed, by a chase into thymidine-containing medium for the indicated times prior to performing iPOND. (B) Quantitation of H4 acetylation levels compared with total H4 in the click reaction samples from three independent experiments. Error bars in all figures are standard deviations. (C,D) Anacardic acid (30 μM) was added to the indicated samples. (E) HU (3 mM) was added to the indicated samples. (F) Cells labeled with EdU were chased into 3 mM HU medium with or without 100 nM FK228 prior to performing iPOND.
In principle, chromatin maturation—as measured by H4K5,K12 deacetylation—could be coupled to fork progression. To test this possibility, we used high concentrations of hydroxyurea (HU) to stall active replisomes and stop DNA synthesis. HU addition stalls the fork effectively in these cells, since the amount of histone capture does not increase appreciably during the HU treatment (Fig. 2E). Deacetylation of newly deposited H4 proceeds at the same rate regardless of whether DNA synthesis is inhibited. Thus, chromatin maturation can be uncoupled from replisome movement.
The histone deacetylase (HDAC) in human cells that catalyzes the deacetylation of H4K5 and K12 is unknown. HDAC1 and HDAC2 associate with CAF-1 (Ahmad et al. 1999), and HDAC3 is required—perhaps in late S phase or G2—to remove H4K5ac (Bhaskara et al. 2010). Indeed, in pulse-chase experiments, we found an enrichment of HDAC1, HDAC2, and HDAC3 near the fork (Fig. 2A), and the selective class I HDAC inhibitor FK228 (Furumai et al. 2002) prevented deacetylation of H4 (Fig. 2F), suggesting that all three of these HDACs are involved.
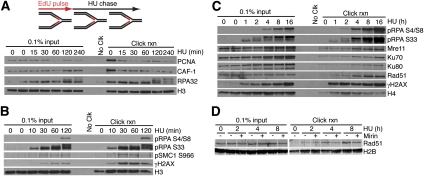
DDR response at stalled replication forks
HU treatment causes DDR activation to stabilize the stalled fork and induce a cell cycle checkpoint. Previous studies suggest that HU-stalled forks remain stable and competent to resume DNA synthesis for several hours; however, eventually, the stalled fork collapses and DSBs are formed (Petermann et al. 2010). To further examine this process, we monitored recruitment and modification of proteins at stalled forks. The amounts of PCNA and CAF-1 that are captured at the stalled fork decrease initially after adding HU to the medium, and then reach a steady state level of between 20% and 30% of that found at an elongating fork (Fig. 3A). This PCNA pattern is likely due to unloading of PCNA from the completed Okazaki fragments. We detected RPA associated with the fork both before and after HU addition (Fig. 3A). The amount of RPA detected remained constant even though RPA accumulates at stalled forks (Cimprich and Cortez 2008). This discrepancy is explained because RPA binds only to the single-stranded, template strand of DNA, which lacks incorporated EdU. Therefore, iPOND detects only the RPA immediately adjacent to the newly synthesized dsDNA (Supplemental Fig. 2).
Figure 3.
iPOND monitors post-translational modifications and recruitment of DDR proteins to stalled and collapsed replication forks. (A–D) Cells were labeled with EdU for 15 min (A) or 10 min (B–D), followed by a chase into HU for the indicated times prior to performing iPOND. (D) HU-treated cells were additionally coincubated with or without the Mre11 inhibitor mirin (100 μM) as indicated.
In these experiments, we noticed that at 120 and 240 min after addition of HU, the electrophoretic mobility of RPA decreased, consistent with phosphorylation (Fig. 3A). RPA S33 phosphorylation could be detected within 10 min of HU addition, and S4/S8 phosphorylation appeared at 2 h (Fig. 3B). DNA-PK catalyzes S4/S8 phosphorylation and ATR catalyzes S33 phosphorylation (Anantha et al. 2007), suggesting that ATR phosphorylates RPA immediately after fork stalling, and DNA-PK phosphorylates RPA only at persistently stalled forks.
H2AX phosphorylation (γH2AX) is often considered a marker for DSBs (Dickey et al. 2009). However, we observed γH2AX at stalled replication forks at even the earliest time points (10 min) after HU addition (Fig. 3B), well before evidence of DSB formation (Petermann et al. 2010). These data prompted us to examine the timing of recruitment of DSB repair proteins. MRE11, KU70, and KU80 exhibited a recruitment profile in which low amounts were observable before the addition of HU, and remained unchanged for 2 h after HU addition (Fig. 3C). However, by 4 h in HU, we detected a significant increase in all of these proteins near the stalled fork (Fig. 3C). RAD51 was first detectable after HU addition, but its levels also increased significantly by 4 h, suggesting that DSBs may form between 2 and 4 h after the fork is stalled. KU70 and KU80 may bind to some of the single-ended breaks, and RAD51 may bind to others.
At DSBs, MRE11-dependent end resection is required to load RAD51 (Mimitou and Symington 2009). At collapsed forks, RAD51 may function to promote recombination-based methods to re-establish the replication fork (Errico and Costanzo 2010). To test whether the loading of RAD51 at stalled forks also requires MRE11, we treated cells with the MRE11 nuclease inhibitor mirin (Dupre et al. 2008). Although the early recruitment of RAD51 occurred independently of MRE11, the late accumulation required MRE11 activity (Fig. 3D), suggesting that end resection promotes this loading. The timing of MRE11 recruitment also correlated with a large increase in RPA S4/S8 phosphorylation (Fig. 3C), which was previously linked to end resection at camptothecin-damaged forks (Sartori et al. 2007).
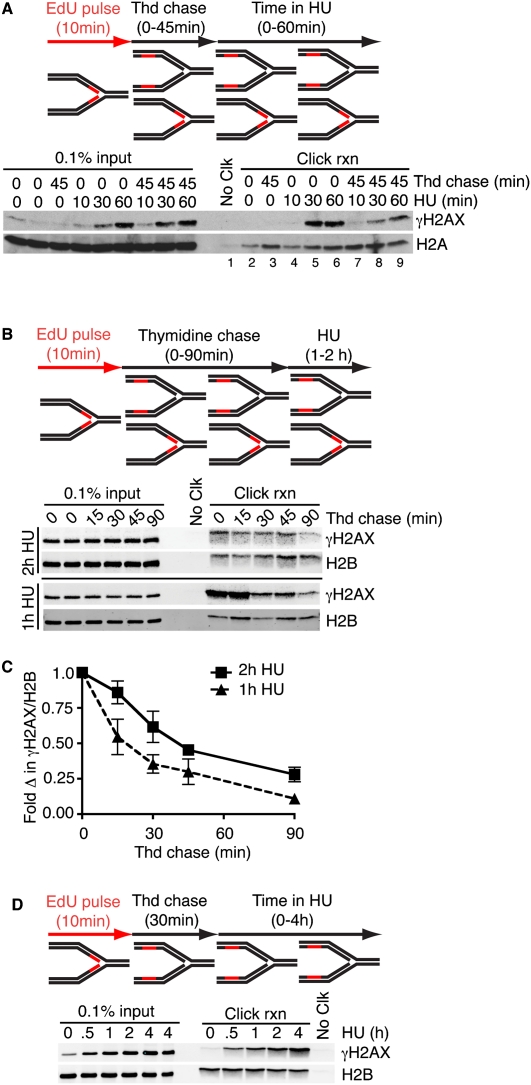
γH2AX spreading from stalled forks before and after fork collapse
We noticed that the rapid phosphorylation of H2AX near the fork saturates within 30 min; however, global levels continue to increase (Fig. 3B, cf. the click rxn lanes and the input lanes). Therefore, we hypothesized that the global increase stems from γH2AX spreading from the stalled fork, as is observed near DSBs (Berkovich et al. 2007; Savic et al. 2009). To test this hypothesis, we first labeled cells with EdU, then chased with thymidine for various lengths of time to extend the distance between the EdU-labeled fragment and the fork, and finally added HU to stall the fork. We again observed maximum γH2AX at the fork 30 min after HU addition; however, the chromatin region distant from the fork contained low but detectable levels of γH2AX that increased when examined at 60 min after HU addition (Fig. 4A, cf. lanes 4–6 and 7–9). A more detailed analysis revealed that the density of γH2AX gradually declined as a function of distance from the stalled fork (Fig. 4B,C). Compared with the saturated density at the fork, the γH2AX density decreased approximately twofold for every 15 min of thymidine chase time when cells were treated with HU for 1 h. By 2 h, we observed increased γH2AX density in all chromatin segments analyzed, suggesting that γH2AX spreading contributes significantly to the global change in γH2AX levels.
Figure 4.
γH2AX spreads from a stalled replication fork. (A–D) Cells labeled with EdU for 10 min were chased into thymidine-containing medium prior to addition of HU, then processed using iPOND. The length of the thymidine and HU treatments is indicated. Quantitation of the click reaction samples in C at the 2-h HU-treated samples is from three independent experiments, and at the 1-h HU-treated samples is from two independent experiments.
To examine the chromatin at a single location distant from the fork, we repeated this experiment holding the thymidine chase time constant at 30 min, and treated with HU for varying times. We observed a steady increase in γH2AX at this distance from the fork (Fig. 4D). Importantly, these results indicate considerable spreading of the γH2AX signal even shortly after fork stalling. Assuming a conservative rate of fork elongation of 1 kb/min, these data imply that, within 1 h of fork stalling, γH2AX spreads to include a large domain containing tens of thousands of base pairs of DNA.
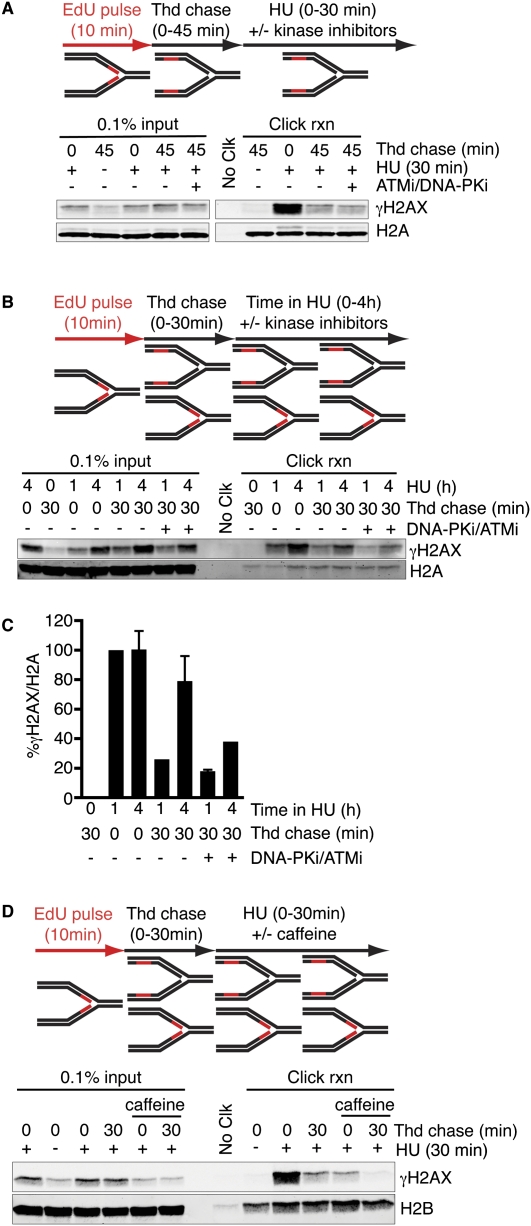
To identify the kinases that phosphorylate H2AX adjacent to the stalled fork and that promote spreading, we used small molecule kinase inhibitors. The selective DNA-PK and ATM inhibitors NU7441 (Leahy et al. 2004) and KU55933 (Hickson et al. 2004) had minimal effects on the spreading or total levels of γH2AX induced by a short (30- to 60-min) HU treatment (Fig. 5A; Supplemental Fig. 3A). However, these inhibitors did significantly reduce γH2AX levels at all chromosomal positions relative to the fork in cells treated with HU for 4 h (Fig. 5B,C; Supplemental Fig. 3B). These results indicate that DNA-PK/ATM contributes to maintenance and spreading of γH2AX at persistently stalled forks. In contrast, treatment with caffeine, which preferentially inhibits ATR (Sarkaria et al. 1999), significantly reduced γH2AX formation and spreading shortly after the fork is stalled (Fig. 5D). These results are consistent with a model in which ATR phosphorylates H2AX at a stalled fork and promotes initial spreading. At later time points, when DSBs likely form at the fork, ATM and DNA-PKcs maintain and further propagate the H2AX phosphorylation (Supplemental Fig. 4).
Figure 5.
Checkpoint kinases propagate H2AX phosphorylation from stalled replication forks. (A–C) Cells labeled with EdU for 10 min were chased into thymidine, followed by treatment with HU. The length of thymidine and HU treatments are indicated. DNA-PK (KU7441, 1 μM) and ATM (KU5593, 10 μM) inhibitors were added at the same time as HU in the indicated samples. (C) Quantitation of the click reaction samples is the average from two independent experiments and is normalized to the 1-h HU treatment. (D) Cells labeled with EdU for 10 min were chased into thymidine for either 0 or 30 min, followed by a 30-min treatment with HU. Caffeine (10 mM) was added at the same time as HU in the indicated samples.
Discussion
Previous studies of the replisome and DDR responses at stalled forks relied largely on immunofluorescent imaging to track protein localization. While useful, immunofluorescence has the significant disadvantages of low resolution and low sensitivity. For example, proteins that exist at only single-copy levels at replication forks cannot be tracked with immunofluorescent imaging. In contrast, iPOND technology has dramatically improved sensitivity, allowing us to detect even proteins such as polymerases. Furthermore, combining iPOND with pulse-chase methods provides high spatial and temporal resolution of protein dynamics. Finally, iPOND also facilitates analysis of post-translational modifications, which is often impossible with immunofluorescent imaging due to poor antibody quality or specificity.
Recently, the Helleday group (Petermann and Helleday 2010) isolated CldU-labeled DNA using an antibody to show that Rad51 is bound to recently synthesized DNA. However, they used a 40-min labeling time, so it is unclear whether this method is sufficiently sensitive or specific to produce high spatial and temporal resolution like iPOND. Also, unlike CldU-IP, iPOND does not require ssDNA to permit antibody access to an antibody epitope, and the biotin–streptavidin purification procedure is compatible with highly stringent conditions (1% SDS and 1 M NaCl), thereby improving specificity.
iPOND is an ensemble methodology. Thus, it provides an average picture of events in different cells at stalled forks throughout the genome. iPOND can be combined with cell synchronization to examine replication and chromatin maturation in early and late replicating genomic regions. In principle, iPOND should be applicable to any process involving DNA synthesis, such as analysis of DNA excision repair.
A disadvantage of iPOND over ChIP methods is the lack of a PCR amplification step. Thus, much larger amounts of input material are necessary to achieve sufficient protein for detection. Fortunately, the covalent coupling of EdU and biotin during the click reaction permits a single-step, highly efficient purification in stringent buffer, salt, and detergent conditions. A significant advantage of iPOND compared with ChIP is its compatibility with unbiased screening approaches. We anticipate coupling iPOND to mass spectrometry to identify all proteins at active and damaged replisomes. Furthermore, mass spectrometry analysis of iPOND-captured histones will facilitate studies of chromatin assembly and maturation.
Chromatin assembly is thought to occur by a stepwise deposition of the core histones, followed by linker histones and changes in post-translational modifications (Probst et al. 2009). Our data confirm this assembly process in vivo in cultured mammalian cells. Furthermore, we found that at least some chromatin maturation processes, such as the removal of acetylation on H4K5 and H4K12, proceed even when decoupled from replisome movement. HDAC1, HDAC2, and HDAC3 are enriched on newly synthesized DNA, and an inhibitor that targets all three of these enzymes prevents H4K5ac and H4K12ac deacetylation. Intriguingly, deacetylation of H4K5ac and H4K12ac occurred at the same rate, but acetyltransferases rapidly reacetylated H4K12, suggesting a specific need for this modification in some chromatin domains.
In the yeast Saccharomyces cerevisiae, H3K56 acetylation is also associated with newly deposited histones during DNA replication, and promotes survival in response to replication stress (Masumoto et al. 2005). We were unable to detect this acetylation mark on newly deposited histones or after HU treatment (data not shown). This observation is consistent with other human cell studies that found low levels of this post-translational modification in total chromatin that further decreased in response to DNA damage (Tjeertes et al. 2009).
Prominent changes in response to replication stress include protein phosphorylation. Importantly, our data indicate that H2AX phosphorylation spreads to a large chromatin domain early in the response to fork stalling. This early phosphorylation is catalyzed by ATR and is unlikely to be due to the processing of the fork into a DSB intermediate. Our data are consistent with previous analyses implicating both ATR-dependent (Ward and Chen 2001) and ATR-independent (Brown and Baltimore 2003; Gilad et al. 2010) H2AX phosphorylating activities in response to fork arrest. Most models of ATR function suggest that it is active only when bound to the ssDNA at the stalled fork through an ATRIP–RPA interaction (Cimprich and Cortez 2008), but our data indicate that ATR helps spread the γH2AX signal. One possibility is that the early spreading of γH2AX is due to looping of the newly synthesized chromatin that brings it into proximity of ATR. Alternatively, ATR may have a method of spreading its signal beyond the immediate ssDNA vicinity, similar to the ability of active ATM to spread along the dsDNA away from the DSB end (You et al. 2007). MDC1 may be involved in such a process (Ichijima et al. 2011; Wang et al. 2011).
Persistent stalling of the fork for longer than 1–2 h causes a switch in the DDR. RPA is hyperphosphorylated on DNA-PK-dependent phosphorylation sites, ATM/DNA-PK catalyzes further γH2AX spreading, and DSB repair proteins like MRE11, KU70/80, and RAD51 accumulate. RAD51 assembly at these persistently stalled forks depends on MRE11 activity, suggesting a requirement for end resection. The end resection may be on the template DNA strand, since we continued to capture EdU-labeled DNA and associated proteins. Resecting the leading strand template would yield a 3′ overhang of newly synthesized DNA, which could be used in recombination-based methods of fork repair and restart (Petermann and Helleday 2010).
Overall, these data provide the first high-resolution, time-dependent analyses of protein dynamics at active, stalled, and collapsed replication forks in mammalian cells. Furthermore, they validate iPOND as a powerful method to study DDRs, chromatin deposition, and chromatin maturation during DNA replication.
Materials and methods
Cell culture
HEK293T cells were cultured in DMEM supplemented with 7.5% FBS. Stable cell lines expressing POLE2-HA and POLE3-HA were generated by retroviral infection and selection in puromycin-containing medium.
Plasmid constructs
POLE2-HA and POLE3-HA retroviral vectors were generated by gateway cloning. pENTR POLE2 and pENTR POLE3 were recombined with pLPCX-GW-HA3X (pDC1127) to generate a C-terminal HA-tagged POLE2 and POLE3 retroviral vectors. pDC1127 was created by subcloning a 3XHA epitope into pLPCX between the Not1 and Cla1 restriction sites, then subcloning the gateway cassette containing attR1, ccdB gene, and attR2 as an EcoRV fragment between EcoR1 and Not1 sites.
iPOND
EdU-labeled sample preparation
HEK 293T cells (∼1.5 × 108 cells per sample) were incubated with 10–12 μM EdU (Vanderbilt Synthesis Core). For pulse-chase experiments with thymidine (Sigma), EdU-labeled cells were washed once with temperature- and pH-equilibrated medium containing 10 μM thymidine to remove the EdU, then chased into 10 μM thymidine. Other chemicals were added to the cell cultures at the following concentrations: HU (3 mM; Sigma), HAT inhibitor anacardic acid (30 μM; Enzo), HDAC inhibitor FK228 (100 nM; kindly provided by Dineo Khabele), Mre11 inhibitor Mirin (100 μM; Sigma), ATM inhibitor (KU55933, 10 μM; AstraZeneca), DNA-PK inhibitor (KU57788, 1 μM; AstraZeneca), and caffeine (10 mM; ICN Biomedicals). DMSO was used as a vehicle control where appropriate.
After labeling, cells were cross-linked in 1% formaldehyde/PBS for 20 min at room temperature, quenched using 0.125 M glycine, and washed three times in PBS. Collected cell pellets were frozen at −80°C, then resuspended in 0.25% Triton-X/PBS to permeabilize. Pellets were washed once with 0.5% BSA/PBS and once with PBS prior to the click reaction.
Click reaction
Cells were incubated in click reaction buffer for 1–2 h at a concentration of 2 × 107 cells per milliliter of click reaction buffer. The click reaction buffer contains Invitrogen's Click-iT cell reaction buffer and cell buffer additive (C10269), 2 mM copper (II) sulfate (CuSO4), and 1 μM photocleavable biotin-azide (Kim et al. 2009) (kindly provided by Ned Porter). DMSO was added instead of biotin-azide to the negative control samples (no clk in all figures). Cell pellets were washed once with 0.5% BSA/PBS and once with PBS.
Cell lysis
Cells were then resuspended in lysis buffer containing 1% SDS, 50 mM Tris (pH 8.0), 1 μg/mL leupeptin, and 1 μg/mL aprotinin. Samples were sonicated (Micro-tip, Misonix 4000 or Fisher Scientific Sonic Dismembrator model 500) using the following settings: 13–16 W, 20-sec constant pulse, and 40- to 59-sec pause for a total of 4–5 min. Samples were centrifuged at 13,200 rpm for 10 min, filtered through a 90-μm nylon mesh, and diluted 1:1 (v/v) with PBS containing 1 μg/mL leupeptin and 1 μg/mL aprotinin prior to purification.
Purification
Streptavidin–agarose beads (Novagen) were washed 1:1 (v/v) twice in lysis buffer and once in PBS. Washed beads were incubated with the samples for 14–20 h at 4°C in the dark. The beads were washed once with lysis buffer, once with 1 M NaCl, and then twice with lysis buffer. Captured proteins were eluted and cross-links were reversed in SDS sample buffer by incubating for 25 min at 95°C. Proteins were resolved on SDS-PAGE and detected by immunoblotting. In most cases, quantitative immunoblotting was performed using the Odyssey infrared imaging system.
Antibodies
Antibodies used were as follows: PCNA (Santa Cruz Biotechnology); CAF-1 p60, RPA32, pRPA32 S4/S8, pRPA32 S33, and pSMC1 S966 (Bethyl Laboratories); FK2 (Calbiochem); RAD51, H2B, H2A, H3, H4, H4K5Ac, KU70, KU80, HDAC1, HDAC2, and HDAC3 (Abcam); γH2AX, H1 (Millipore); MRE11 (Genetex); H4K12Ac and H4K20me1 (Active Motif); and anti-HA (Covance).
Determination of DNA fragment size
To determine DNA fragment size, 5 μL of pre- and post-sonication samples were incubated at 65°C to reverse the DNA–protein cross-links, then incubated with RNaseA and proteinase K. DNA samples were resolved on a 1.5% agarose gel, stained with ethidium bromide, and visualized under UV light. DNA fragment sizes ranged between 100–300 bp. It should be noted that we determined that the CuSO4 in the click reaction catalyzes cleavage of the phosphodiester bond and assists in generating the small fragment size.
Acknowledgments
We thank Drs. Ned Porter, Keri Tallman, Dineo Khabele, Janel McLean, Kathy Gould, Mahesh Chandrasekharan, Simona Codreanu, Daniel Liebler, and Larry Marnett for supplying reagents and advice. This work was supported by NIH grant R01CA136933 to D.C. Additional support was provided by the Vanderbilt-Ingram Cancer Center (CA06485) and the Ingram Charitable Fund. B.M.S. is supported by a Department of Defense Breast Cancer Research Program predoctoral fellowship (W81XWH-10-1-0226). F.B.C is supported by the Molecular Toxicology training grant (T32 CA09582).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2053211.
Supplemental material is available for this article.
References
- Ahmad A, Takami Y, Nakayama T 1999. WD repeats of the p48 subunit of chicken chromatin assembly factor-1 required for in vitro interaction with chicken histone deacetylase-2. J Biol Chem 274: 16646–16653 [DOI] [PubMed] [Google Scholar]
- Anantha RW, Vassin VM, Borowiec JA 2007. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem 282: 35910–35923 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2008. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc 3: 915–922 [DOI] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. 2010. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev 17: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM 2009. H2AX: functional roles and potential applications. Chromosoma 118: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 4: 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V 2010. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep 11: 270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, et al. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 62: 4916–4921 [PubMed] [Google Scholar]
- Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, Brown EJ 2010. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res 70: 9693–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ 2007. The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Herrick J, Bensimon A 2008. Global regulation of genome duplication in eukaryotes: an overview from the epifluorescence microscope. Chromosoma 117: 243–260 [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH 2011. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25: 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Tallman KA, Liebler DC, Porter NA 2009. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol Cell Proteomics 8: 2080–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14: 6083–6087 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Shen X 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses JE, Moorhouse AD 2007. The growing applications of click chemistry. Chem Soc Rev 36: 1249–1262 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25: 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D, Truman AW, Kron SJ, Cote J 2010. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res 16: 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Haber JE 1988. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol 8: 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59: 4375–4382 [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH 2009. Formation of dynamic γ-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell 34: 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci 92: 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G 1999. Duplication and maintenance of heterochromatin domains. J Cell Biol 147: 1153–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP 2009. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J 28: 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR 2010. Modifying chromatin architecture during the response to DNA breakage. Crit Rev Biochem Mol Biol 45: 2–13 [DOI] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J 2011. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 193: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276: 47759–47762 [DOI] [PubMed] [Google Scholar]
- Worcel A, Han S, Wong ML 1978. Assembly of newly replicated chromatin. Cell 15: 969–977 [DOI] [PubMed] [Google Scholar]
- You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T 2007. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol 9: 1311–1318 [DOI] [PubMed] [Google Scholar]