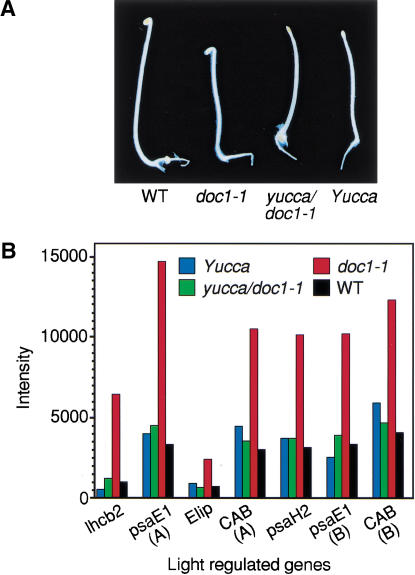
Figure 2.
Suppression of overexpression of light-regulated genes in dark-grown doc1 by auxin overproducing yucca. (A) Four-day-old dark-grown wild type, doc-1, yucca, and doc1-1 yucca double mutant. (B) Expression levels of some light-regulated genes in dark-grown seedlings analysed by microarray. The Affymetrix accession nos. for CAB(A), CAB(B), psaE1(A), psaE1(B), lhcb2, Elip, and psaH2 are 16004-s-at, 13213-s-at, 18088-i-at, 18089-r-at, 15153-at, 16637-s-at, and 18081-at, respectively. The CAB(A) oligos were designed based on the genomic sequence of the CAB gene (GenBank AL049655), and CAB(B) oligos were based on the mRNA sequence of the CAB gene (GenBank X56062). Both sets of oligos for the CAB gene may also pull out other homologous genes such as CAB2 in the experiments shown here. Oligos for both psaE1(A) and psaE1(B) were designed based on the mRNA sequence of the photosystem-I subunit IV precursor (GenBank AJ245908). Both sets of psaE1 oligos were designed for detecting the same gene, but neither of them is perfect based on Affymetrix rules for oligo design. There are <15 oligos in the set for psaE1(A), whereas some rules for oligo design were dropped for the psaE1(B). Microarray experiments were performed using instructions provided by Affymetrix.