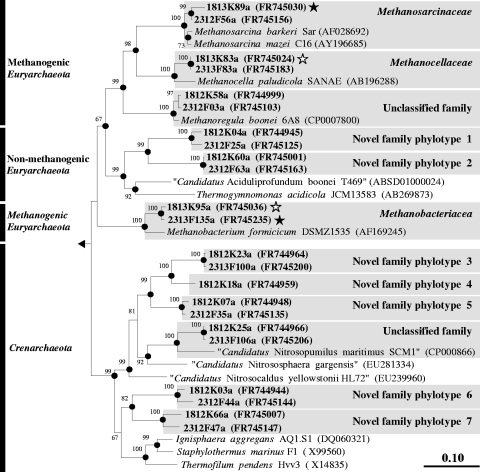
Fig. 6.
Phylogenic neighbor-joining tree of representative family-level archaeal 16S rRNA gene sequences retrieved from formate and CO2 treatments and reference sequences. Values next to the branches represent the percentages of replicate trees (>50%) in which the associated taxa clustered together in the bootstrap test (10,000 bootstraps). Dots at nodes indicate the confirmation of tree topology by maximum-likelihood and maximum-parsimony calculations with the same data set. Quotation marks indicate provisional taxa (J. P. Euzéby, List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.cict.fr/number.html]). Bar indicates a 0.1 estimated change per amino acid. E. coli (X80725) was used as an outgroup. Clones are identified, in order, by the number of days postsupplementation (18 or 23) the type of treatment (12K, 12CO2 treatment; 12F, [12C]formate treatment; 13K, 13CO2 treatment; 13F, [13C]formate treatment) and clone number, followed by “a” to indicate archaeal sequence. Accession numbers are given in parentheses. Symbols: filled stars, labeled phylotypes; empty stars, marginally labeled phylotypes.