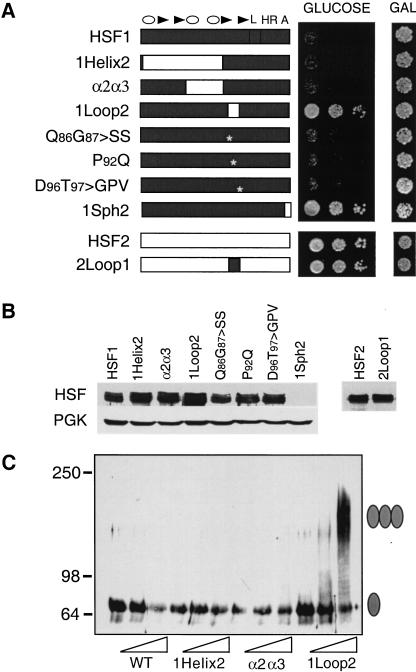
Figure 5.
The HSF1 loop contributes to suppression of HSF1 activity in yeast. (A) Complementation assay for the ability of mouse HSF1, HSF2, and chimeras to substitute for yeast (heat-shock transcription factor) HSF. Cells harboring a chromosomal disruption of yeast hsf1 and bearing a plasmid with (heat stress transcription factor) HSF under the control of the GAL1 promoter were transformed with another plasmid expressing the indicated mouse HSF from a constitutive promoter. Transformants were diluted serially (from left to right, OD650, 0.1, 0.01, 0.001) and spotted onto glucose- or galactose-containing medium, incubated for 3 d at 30°C and photographed. (B) Western analysis for expression of mouse HSF isoforms in yeast using antibodies specific to HSF1 or HSF2. Levels of PGK are shown for normalization of loading. The 1Sph2 isoform is not detected with the HSF1 antibody because the primary epitopes recognized by this antibody map to the carboxy-terminal regions of the protein, which have been replaced by HSF2 in the chimera. (C) Crosslinking of HSF reveals that the HSF1 loop suppresses trimerization. Whole-cell extracts were prepared from yeast cells expressing the indicated forms of mouse HSF and subjected to in vitro cross-linking with 0, 0.5, or 2 mM EGS (indicated by wedge). HSF proteins were detected by immunoblotting with HSF1-specific antibody. The monomeric and trimeric HSF species are depicted with a single or triple oval, respectively.