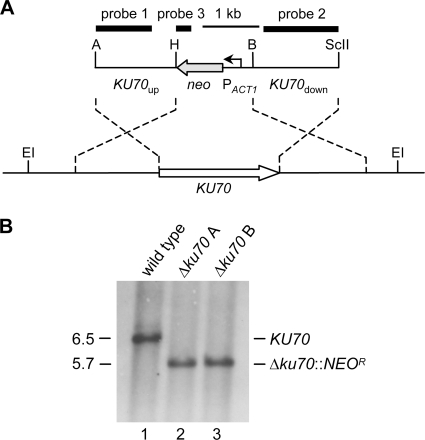
Fig. 2.
Construction of A. benhamiae Δku70 mutants. (A) Structure of the deletion cassette from plasmid pAbenKU70M2 (top), containing the neomycin resistance marker (NEOR), and genomic structure of the KU70 locus in wild-type A. benhamiae Lau2354-2 (bottom). The KU70 coding region is represented by the white arrow, and the upstream and downstream regions are represented by the solid lines. The neomycin resistance gene (neo) is shown as a gray arrow, and the A. benhamiae ACT1 promoter (PACT1) as a bent arrow. The probes which were used for Southern analysis of the transformants are indicated by the black bars. Only the following relevant restriction sites are given: A, ApaI; B, BamHI; EI, EcoRI; H, HindIII; ScII, SacII. (B) Southern hybridization of EcoRI-digested genomic DNA of the wild-type strain A. benhamiae Lau2354-2 (lane 1) and Δku70 mutants AbenKU70M1A (lane 2) and AbenKU70M1B (lane 3) with KU70-specific probe 1. The sizes of the hybridizing fragments (in kilobases) are given on the left, and their identities on the right.