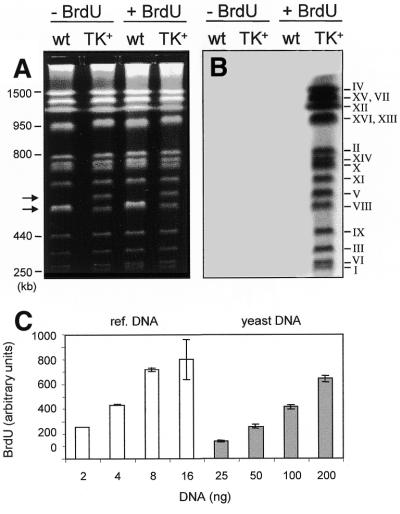
Figure 1.
Incorporation of BrdU into chromosomes of GPD–TK7× yeast cells. (A) Exponential cultures of E001 (wild-type) and E1000 (TK+) cells were grown for 2 h at 25°C in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 400 µg/ml BrdU. Chromosomal DNA samples were prepared and separated by PFGE as described in Materials and Methods (except that the 60 s pulses were for 8 h only) and stained with ethidium bromide. The position of molecular weight markers is shown on the left. Arrows point to chromosome V, 576 and 611 kb (due to the insertion of seven GPD–TK copies) in the wild-type and TK+ strains, respectively. (B) The gel shown in (A) was transferred to a nitrocellulose membrane and BrdU was detected using a monoclonal antibody against BrdU and a fluorescently labeled secondary antibody. After FluorImager scanning a signal is detected only in TK+ cells grown in the presence of BrdU. (C) Relative BrdU levels in known amounts of fully substituted reference DNA (left) and total yeast DNA after one round of DNA replication with 400 µg/ml BrdU (right). Purified reference and test DNAs were denatured by boiling, spotted on a membrane and BrdU quantitated using anti-BrdU and fluorescent secondary antibodies. After correcting for one-strand synthesis, the ratio of BrdU versus dT incorporated in vivo is 0.1. Mean values ± SD are shown for three independent experiments.