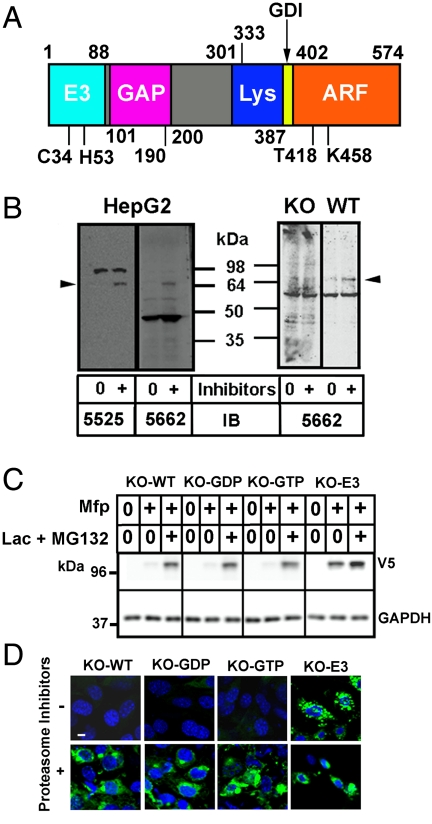
Fig. 1.
ARD1, a multifunctional protein. (A) Functional domains of 574 amino acid human ARD1 with C-terminal (402–574) ARF sequence. ARD1 contains an E3 ubiquitin ligase motif (positions 31–75) with critical C34 and H53, which were replaced with alanine in E3-inactive mutants. GAP function was assigned to amino acid 101–190, with amino acid 190–333 additionally required for domain interactions that result in GTP hydrolysis. A region required for lysosomal targeting (amino acid 301–402) includes amino acid 387–402 corresponding to the N-terminal amphipathic helix of an ARF molecule, which can act as a GDP-dissociation inhibitior (GDI). Mutations in the ARF region (402–574) that abolish GTP binding (T418N) or GTPase activity (K458I) are shown. (B) Effect of proteasome inhibitors on ARD1 content in HepG2 cells and MEFs. HepG2 cells were incubated 18 h without (0) or with (+) lactacystin and MG132, before preparation of lysates and Western blotting of proteins (40 µg) with ARD1 antibodies 5525 or 5662. MEFs lacking ARD1 (KO) or from wild-type mice (WT) were incubated ca. 18 h without or with proteasome inhibitors before Western blotting using ARD1 antibodies 5662. Arrowhead, endogenous ARD1 protein. (C) ARD1-/- MEF lines stably transfected with indicated GFP–ARD1-V5/His constructs (KO-WT, KO-GDP, KO-GTP, KO-E3) were incubated 18 h without (0) or with (+) Mfp or Mfp plus lactacystin and MG132 before Western blotting (40 µg proteins) with antibodies against V5 or GAPDH. Results were replicated in three experiments. (D) MEFs were incubated with Mfp without or with proteasome inhibitors (as in C) before fixation and preparation for inspection of EGFP–ARD1 by confocal immunofluorescence microscopy.