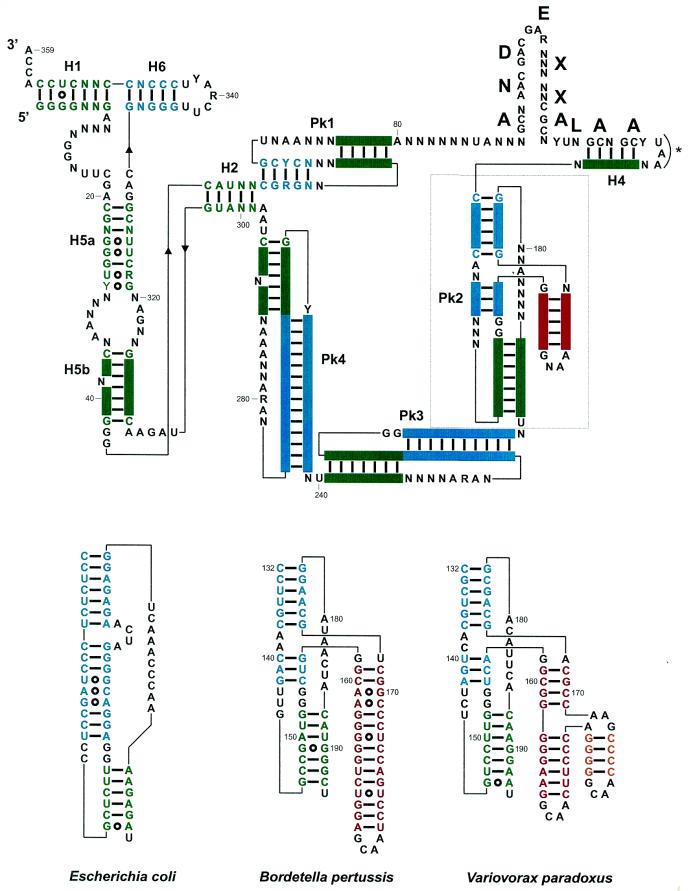
Figure 2.
(Top) Consensus secondary structure for tmRNAs from the β-proteobacteria, with the color code used in Figure 1. Structural domains are H1–H6 for the RNA helices, and Pk1–Pk4 for the pseudoknots. A predicted amino acid sequence consensus is shown above the coding sequence. The bold letters correspond to the encoded amino acids, with X corresponding to non-conserved amino acids. Pk2 is boxed, and its structural insertion, specific of tmRNAs from the β-proteobacteria, is in red. The stop codon is marked by an asterisk. (Bottom) Phylogenetically supported secondary structures of the additional domain (in red and yellow) within Pk2 from representative β-proteobacteria, V.paradoxus and B.pertussis compared to E.coli. Notice the incremental intricacy of the structural insertion. Solid bars between bases represent the Watson–Crick base pairs and open circles the non-Watson–Crick pairs. The two invariant G residues (positions 153 and 154) at the junction between the helices forming Pk2 require some discussion. They could form pairs with positions 143 and 144 (as positions 151–152 and 163–164 do in E.coli) and the drawings shown in Figure 3 would tend to support that view. However, two points prevent a definite conclusion; first, the bases vary on only one strand and, secondly, the relative lengths of the helices forming Pk2 vary so that one cannot derive a unique alignment between the β-proteobacteria and E.coli. Thus, they are shown as unpaired.