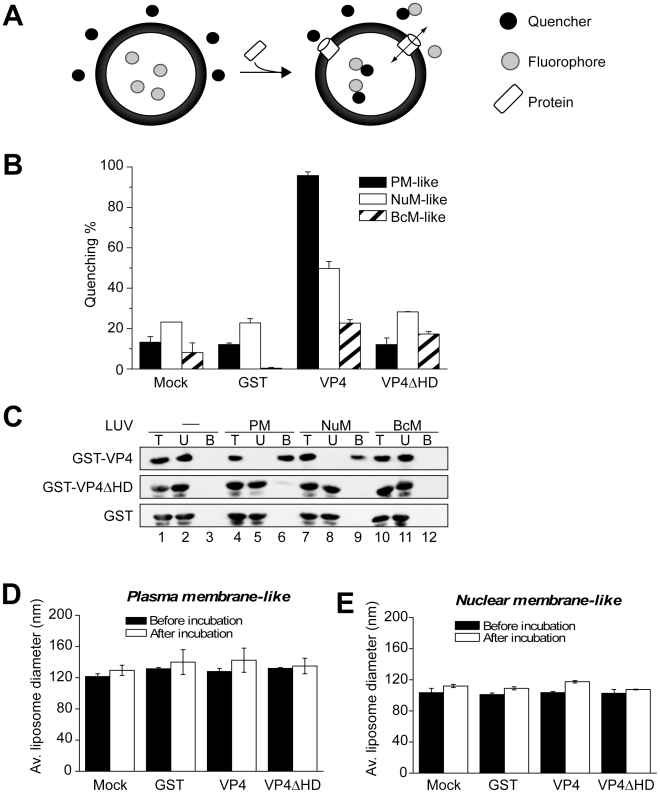
Figure 5. VP4 permeabilization of liposomes.
(A) Scheme showing the liposome disruption assay employed. Membrane disruption was examined by encapsulating [Tb(DPA)3- 3] fluorophore into LUVs (large unilamellar vesicles). When these LUVs were resuspended in a solution containing EDTA (quencher), protein mediated membrane disruption was monitored by the quenching of [Tb(DPA)3- 3] emission as the encapsulated molecules were released, and terbium ions were chelated by EDTA. (B) VP4 disrupts mammalian plasma (PM-like) and nuclear membrane-like (NuM-like) LUVs. LUVs mimicking the lipid compositions of bacterial inner membrane (BcM-like), and mammalian plasma and nuclear membranes were prepared to examine the membrane disruption activity of GST-VP4 and GST-VP4ΔHD. GST was used as a control. Mock LUVs were incubated in absence of any protein. Liposome disruption was evaluated using LUVs prepared with selected lipid compositions, and the percentage of fluorophore quenched is indicated. Each data point shows the average of at least two independent measurements with error bars representing standard deviations. (C) Flotation of proteins on sucrose gradients after incubation either without (-, lanes 1–3) or with PM-like (lanes 4–6), NuM-like (lanes 7–9), or BcM-like (lanes 10–12) LUVs to separate unbound (U; lanes 2, 5, 8 and 11) and bound (B; lanes 3, 6, 9 and 12) fractions. Proteins were resolved by SDS-PAGE and immunoblotting with anti-GST antibody. (D) Average diameter of PM-like LUVs before and after 30 min incubation with the indicated proteins as determined by dynamic light scattering. Each data point shows the average of at least two independent measurements with error bars representing standard deviation. (E) Average diameter of NuM-like LUVs determined similarly to D.