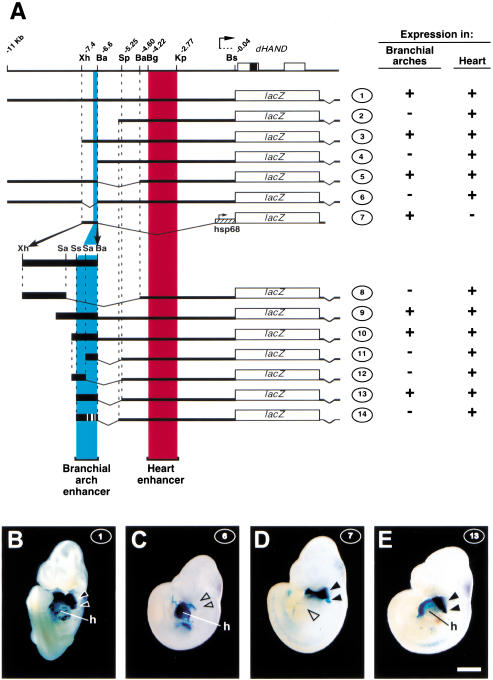
Figure 1.
Identification of a branchial arch-specific enhancer. (A) Reporter constructs used in this study. The genomic organization of dHAND is depicted at the top; coding regions (boxes), relevant restriction sites, and their distance in kilobases from the start of the coding region are indicated. The lacZ reporter constructs used in this study are schematized underneath; thick black lines and boxes represent dHAND genomic sequences. Construct 2 extends to −5.49 kb. The Sau3A, SspI, and Sau3A sites used in generating constructs 8 and 11–14 are located 316, 203, and 121 bp, respectively, upstream of the BamHI site at −6.6 kb. The 5′ ends of constructs 9 and 10 are 413 and 196 bp, respectively, upstream of this BamHI site. Presence or absence of β-galactosidase expression driven by these constructs at E10.5 in branchial arches 1 and 2 and in the heart, respectively, is indicated on the right. (Ba) BamHI; (Bg) BglII; (Bs) BssHII; (Kp) KpnI; (Sa) Sau3A; (Sp) SpeI; (Ss) SspI; (Xh) XhoI. (B–E) Ventral/lateral views of E10.5, Xgal-stained F0 embryos transgenic for constructs 1, 6, 7, and 13, as indicated at upper right of panels. At least three F0 transgenic embryos were examined for each construct. B shows β-galactosidase activity in heart (h) and the first two branchial arches (black arrowheads). (C) Expression in heart but not in branchial arches (open arrowheads). (D) Expression in branchial arches (filled arrowheads) but not in heart (open arrowhead). (E) Expression in heart and branchial arches. Bar, 1 mm.