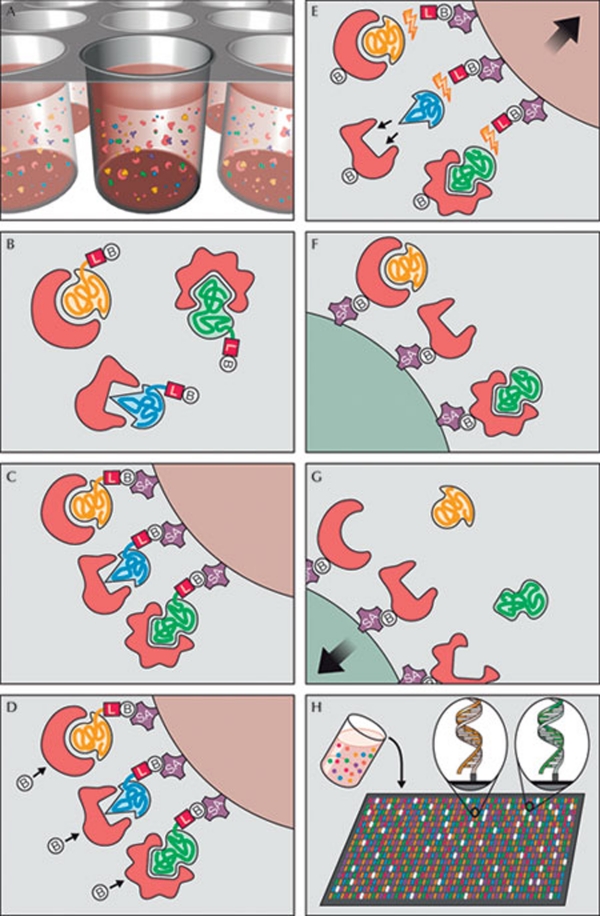
Figure 2.
Principle of multiplex SOMAmer affinity assay, a new aptamer-based proteomic technology for biomarker discovery capable of simultaneously measuring thousands of proteins from small sample volumes. (A) Aptamers (SOMAmers)—short stretches of nucleic acids that form protein-binding three-dimensional structures—and samples are mixed in 96-microwell plates and allowed to bind. Cognate and non-cognate SOMAmer-target protein complexes form. Free SOMAmer and protein are also present. (B) SOMAmer molecules (yellow, blue, and green) have unique shapes selected to bind to a specific protein. SOMAmers contain biotin (B), a photo-cleavable linker (L) and a fluorescent tag at the 5´ end. Most SOMAmers (yellow and green) bind to cognate proteins (red), but some (shown in blue) form non-cognate complexes. (C) SOMAmers are captured onto a bead coated with streptavidin (SA), which binds to biotin. Uncomplexed proteins are washed away. (D) Proteins are tagged with NHS-biotin. (E) UV light (hν) cleaves the linker and SOMAmers are released from beads, leaving biotin on the bead. Samples are challenged with an anionic competitor (dextran sulphate). Non-cognate complexes (blue SOMAmer) preferentially dissociate. (F) SOMAmer–protein complexes are captured onto new avidin-coated beads by protein biotin tag. Free SOMAmers are washed away. (G) SOMAmers are released from complexes into a high-pH solution. (H) The remaining SOMAmers are quantified by hybridization to microarray containing single-stranded DNA probes complementary to SOMAmer DNA sequence, which form a double-stranded helix. Hybridized SOMAmers are detected by fluorescent tags when the array is scanned. Figure credit: Gold et al (2010). http://creativecommons.org/licenses/by/2.5