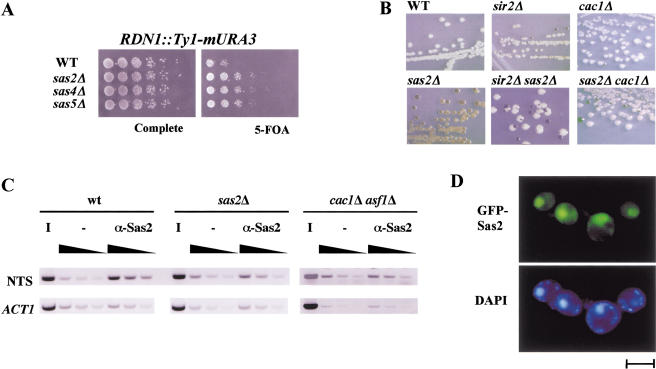
Figure 2.
SAS-I was involved in rDNA silencing. (A) The deletion of SAS2, SAS4, or SAS5 increased repression of URA3 inserted in the rDNA locus. Silencing of URA3 was measured as growth on 5-FOA-containing medium. The strains used were AEY1778, AEY2279, AEY2360, and AEY2361 (from top to bottom). (B) Increased silencing of MET15 by sas2Δ in rDNA required SIR2. Strains were assayed for MET15 activity by streaking them on lead indicator medium. The strains used were AEY1201 (WT), AEY1202 (sir2Δ), AEY1195 (sas2Δ), AEY1978 (sas2Δ sir2Δ), AEY1755 (cac1Δ), and AEY2487 (sas2Δ cac1Δ). (C) Sas2 was physically associated with rDNA sequences. Chromatin immunoprecipitations (CHIPs) were performed with extracts from a wild type (AEY1), a sas2Δ strain (AEY269), and a cac1Δ asf11Δ (AEY2451) strain using α-Sas2 or no antibody (−). PCR was performed with increasing amounts of precipitate (black triangle) using primers specific to the nontranscribed spacer of RDN1 (NTS) and ACT1. The inverse image of PCR products resolved on ethidium bromide-stained 1% agarose gels is shown. (D) GFP-tagged Sas2 localized to the nucleus. The sas2Δ strain AEY474 was transformed with pAE94. Bar, 5 μm.