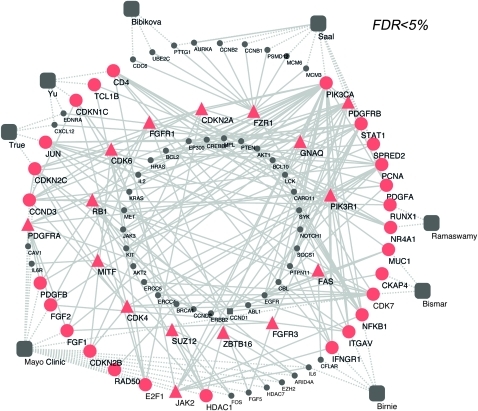
Figure 2.
Combined network of prioritized signature genes and cancer proteins derived from single protein analysis of network (SPAN) protein interaction analysis conducted over each expression signature. Prostate cancer gene signatures of poor prognosis (large grey squares) were evaluated for their protein–protein interaction connectivity to the Sanger cancer genes curated by the Wellcome Trust Cancer Gene Atlas through SPAN methodology. Squares represent prostate cancer gene signatures, circles indicate network genes, and triangles indicate Wellcome Trust Sanger cancer genes. Red indicates statistically significant proteins (false discovery rate (FDR) <5%) with at least two interacting partners, and grey indicates non-prioritized proteins. Nodes on the outer circle indicate prostate cancer signature genes, and nodes in the innermost circle indicate proteins contributing to prioritize the statistically significant ones but for which the FDR >5%. Dashed lines indicate linkages between signature genes and their respective signatures, and solid lines indicate a protein interaction.