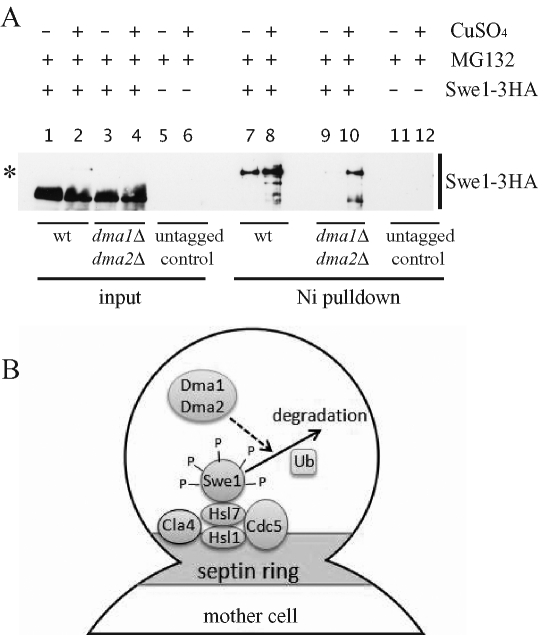
FIGURE 8:
Swe1 ubiquitylation is partially dependent on the Dma proteins. (A) Exponentially growing cultures of wild-type SWE1-3HA (yRF957) and dma1Δ dma2Δ SWE1-3HA (yRF959) cells, as well as of wild-type cells not carrying the SWE1-3HA fusion (yRF1138; untagged control), all containing a high–copy number plasmid with the CUP1–6xHIS-UBI4 construct, were incubated for 3 h in the presence of the proteasome inhibitor MG132 and with (lanes 2, 4, 6, 8, 10, and 12) or without (lanes 1, 3, 5, 7, 9, and 11) 250 μM CuSO4, which induces overproduction of 6xHIS-ubiquitin. Cells were then lysed, and aliquots of the lysates were incubated with Ni-NTA beads to purify the proteins bound by 6xHIS-ubiquitin (Ni pulldown). Bead eluates were subjected to SDS–PAGE electrophoresis and Western blot analysis with anti-HA antibodies. Equivalent aliquots of the lysates were also loaded on the gel (input). The asterisk marks Swe1-ubiquitylated bands. (B) A model for Swe1 down-regulation by the Dma proteins. Swe1 is recruited by the Hsl1/Hsl7 complex to the septin ring, where it interacts with the protein kinases Cla4 and Cdc5 and undergoes multiple phosphorylation events (P). Phosphorylated Swe1 is then likely polyubiquitylated, followed by degradation. The Dma1 and Dma2 proteins partially trigger this degradation by acting after recruitment of Swe1 to the bud neck and its subsequent phosphorylation, possibly by controlling, directly or indirectly, Swe1 ubiquitylation.