Fig 1.
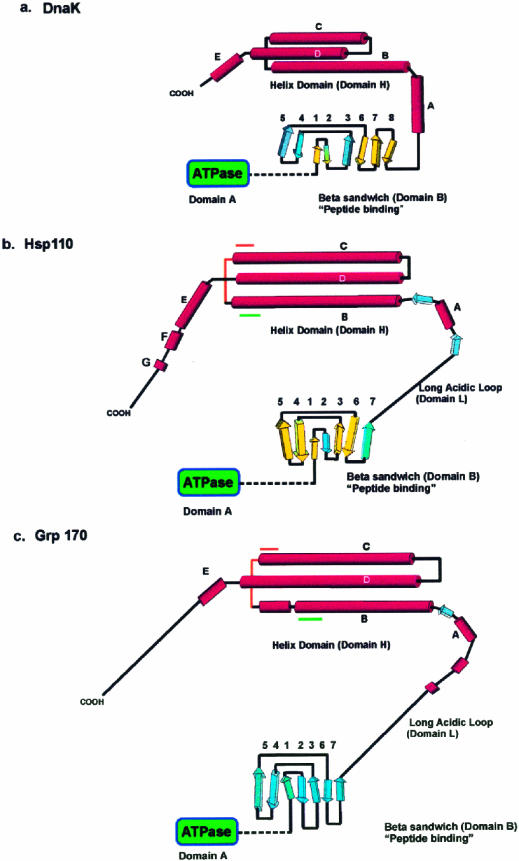
Models for the fold of Hsp110 and Grp170 based on the structure of DnaK (modified from Oh et al 1999). (A) The secondary structure and fold of the DnaK peptide-binding domains based on the x-ray crystallographic structure reported by Zhu et al (1996). The predicted secondary structure and proposed fold for (B) Hsp110 and (C) Grp170, beginning at residue 390, are diagrammatically represented. The domains (A, B, L, and H), as defined in the text, are indicated on the diagram. The β-strands in DnaK and Hsp110 β-domains, sharing both structural (PHD) and significant sequence similarity by Matchbox alignment (Depiereux et al 1997), appear in yellow. Although there is sequence similarity between the β-domains of Hsp110 and Grp170, there is not enough structural and sequence similarity to allow us to identify homologous strands. The predicted helical segments in Hsp110 and Grp170 are shown arranged in a helix-turn-helix structure, as suggested by the helical domain in DnaK. The positions of the conserved sequence motifs (Magic and TedWylee) are indicated by green and orange lines
