Fig 4.
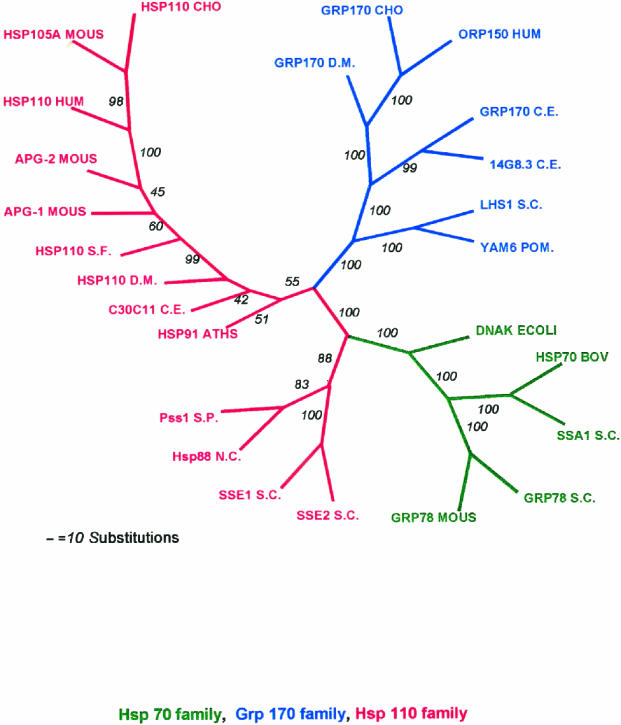
Phylogenetic tree for Hsp70 superfamily. Eleven members of the Hsp110/SSE family and 9 members of the Grp170 family were aligned with 5 members of the Hsp70/DnaK family using the CLUSTAL W program, and the alignments were analyzed by programs in PHYLIP (Phylogeny Inference Package) version 3.57c. The alignments were bootstrapped with SEQBOOT to generate 100 sets of alignment data. The data sets were used to generate 100 phylogenetic trees by use of the POTOPARS (maximum parsimony) program. A consensus tree was determined by use of the program CONSENSE. The tree is displayed as unrooted, with the italicized numbers indicating the number of trees in which the consensus bifurcation distal to the number shown was represented in the total set of 100 trees. Abbreviations and sequences used are the same as in Tables 2 and 3, except the following additional sequences were used in the tree: HSP110CHO (Q60446) Chinese hamster, APG-1 MOUS (D49482) mouse, APG-2 MOUS (Q61316) mouse, YAM6 S. POM (Q10061) S pombe, 14G8.3 C.E. (CAA91809) C elegans, SSA1 S.C. (6319314), S cerevisiae Grp78S.C.; KAR2 (6322426) S cerevisiae, HSP70BOV (P34933) bovine
