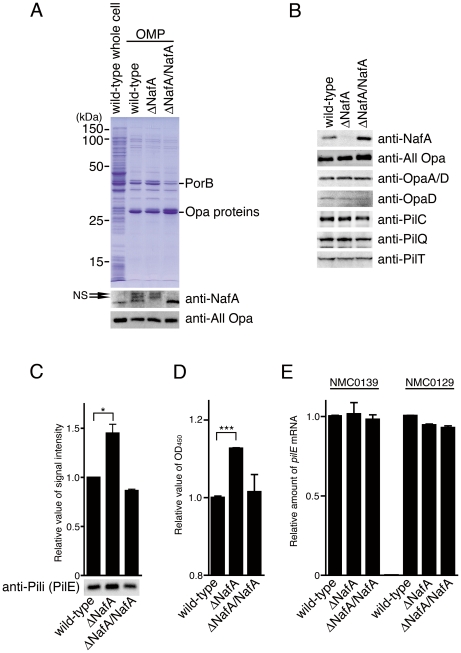
Figure 3. The ΔNafA strain expresses more PilE protein than the wild-type strain.
Bacterial outer membrane proteins (A) and whole cell lysate samples (B) were prepared from strains indicated and separated by SDS-PAGE. A. Expression profiles of outer membrane protein were detected by Coomassie Brilliant Blue staining. PorB and Opa proteins were confirmed by MALDI-TOFMS. The lower panel shows the expression of NafA and Opa proteins in the outer membrane fraction, as detected by western blot analysis using anti-NafA peptide C antibody and anti-All Opa antibody, respectively. Arrows indicate non-specific bands (NS). B. Detection of NafA, Opa proteins and pilus components from whole bacterial lysate samples by western blot analysis. Equal amounts of whole cell lysates were loaded in each western blot analysis. C. The western blot analysis was performed as panel B. The signal intensities were analyzed using ImageJ software and relative value were presented when value of the wild-type strain was set as 1.0. D. Amounts of PilE produced by indicated strains were compared by whole cell ELISA using anti-pili (which mainly recognizes PilE, see Experimental procedures). The histogram shows the relative PilE expression levels of bacterial strains, OD450 value of the wild-type strain was set as 1.0. The error bars in C and D represent standard error of mean (SEM) from triplicate experiments. Statistically significant difference is indicated with single asterisk (P<0.05) or triple asterisks (P<0.001). E. Level of pilE mRNA were compared by quantitative RT-PCR. The histogram shows relative pilE mRNA levels normalized to the housekeeping genes, NMC0139 (50S ribosomal protein rplP; left three bars) or NMC0129 (30S ribosomal protein rpsJ; right three bars), and calculated in arbitrary units set to a value of 1 for the wild-type strain. The experiments were repeated using samples generated from two separate cDNA synthesises. Also, cDNA synthesized from a separate RNA preparation gave similar results (data not shown).