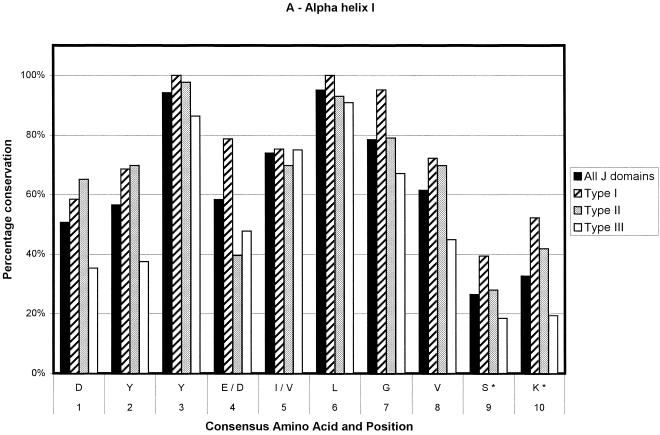
Fig 2.
Bar chart representations of the percentage conservation of each amino acid in the consensus sequences derived from an alignment of all J domains and the J domains of type I, II, and III DnaJ-like proteins. The data have been separated into helix I (a), helix II (b), the loop region (c), helix III (d), and helix IV (e). The single letter amino acid code has been used. The different consensus sequences were derived by determining the most highly conserved amino acid based on percentage identity at each position in all 223 J domain sequences and the J domains from 91 type I, 43 type II, and 89 type III DnaJ-like proteins. Only the consensus residues derived from the alignment of all J domain sequences are given. The positions where there is variation among the 4 derived consensus sequences are indicated with an asterisk and are detailed in Table 1