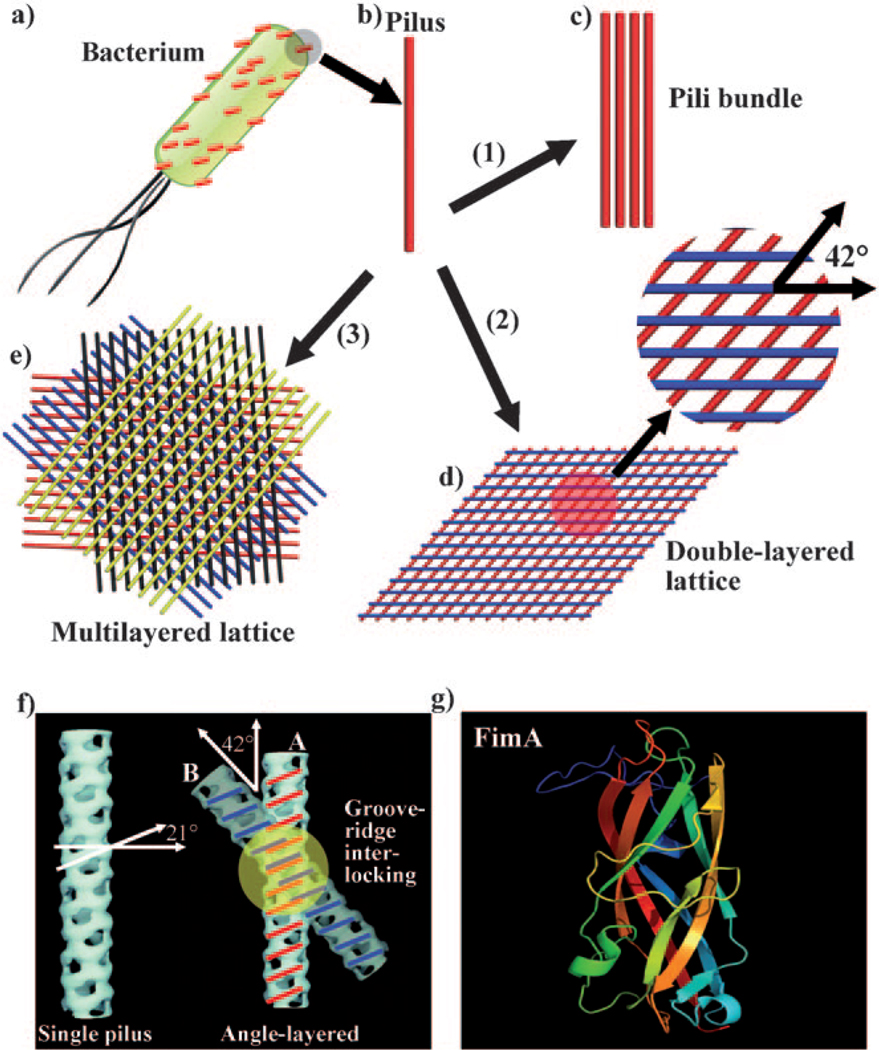
Figure 1.
Self-assembly of pili into 1D bundles, 2D lattices, and 3D lattices. a) Illustration of a bacterial cell with many pili protruding from its surface. b) An individual pilus. c) Pili bundles. d) Double-layer pili lattices. The two neighboring layers of pili have a fixed angle of 42°. e) Four-layer pili lattices. A red layer of parallel pili was formed, with subsequent assembly of a blue layer of pili on top of the red layer with a twist angle of 42°. After that, a black and a yellow layer are deposited in turn on top of the blue layer and black layer, with the neighboring layer twisted at the same angle of 42°, resulting in the formation of a four-layer pili lattice. f) Molecular recognition between two pili particles. A single pilus has a set of alternating ridges and grooves with a pitch angle of 21°. When the crossing angle between pili A and B is 42°, the ridges of pilus A can make a reasonable interlocking fit into the corresponding grooves of pilus B. g) The 3D structure of the subunit of pili FimA (PDB ID: 2JTY). Inducing agents include: 1) more than 160 mm hexamethylenediamine; 2) 80 mm hexamethylenediamine; and 3) 80 mm pimelic acid or 1,3-propanedisulfonic acid. A high-resolution version of (e) is shown in Figure S3 in the Supporting Information.