Erratum to: Arch Virol (2010) 155:37–46 DOI 10.1007/s00705-009-0548-9
Figure 1 (and figure legend) on page 41 and the first paragraph on page 42 should be replaced with the following figure (and figure legend) and text, respectively:
Fig. 1.
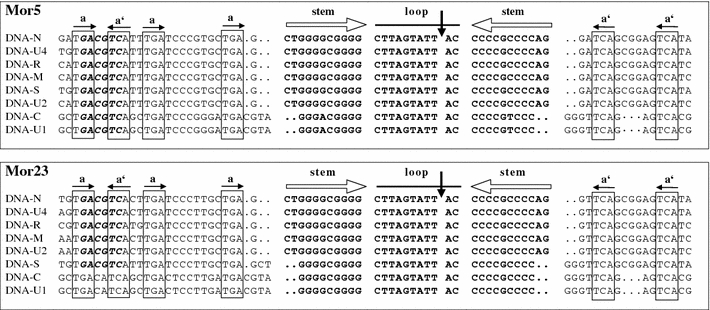
Alignment of the replication origin sequences of the eight DNAs of the isolates Mor5 and Mor23 illustrating the extent of sequence conservation in the stem-loop common region (CR-SL). Inverted repeat sequences (open horizontal arrows) potentially forming a stem-loop structure are in bold. A vertical arrow indicates the ‘origin of replication’, which is the position of cleavage by the master Rep protein [44]. Conserved iteron-like sequences (a) and their respective inversions (a′) possibly acting as recognition or binding sites for the master Rep protein are boxed and indicated by horizontal solid arrows. Dots indicate gaps included to maximise alignments. The AatII site (GACGTC) conserved in many nanoviruses [17] but absent from DNA-C and -U1 of Mor23 is shown in bold italics
The noncoding regions (NCRs) of all eight DNAs of each isolate shared a highly conserved, inverted repeat sequence predicted to form a stem-loop structure and characteristic of nanovirus DNAs [7, 26, 28, 39, 43]. An alignment of these conserved stem-loop and flanking iteron-like sequences of the Mor5 and Mor23 DNAs is shown in Fig. 1. The stem-loop sequences [nt –18 (or –20) to +13 (or +11)] of Mor5 are identical to those of FBNSV-Eth (not shown). These two isolates are also indistinguishable in the component (DNA-C and -U1)-specific differences (A/G and C/T at nt positions –15 and +8, respectively). The nonanucleotide sequence TAGTATT↓AC, which encompasses the origin of replication [44], is perfectly conserved in all the DNAs of both isolates. Moreover, the domains flanking the stem-loop region are largely conserved in all the DNAs. The aforementioned component (DNA-C and -U1)-specific differences are absent from the stem-loop sequences of Mor23. However, Mor23 has other component-specific differences not only in the stem-loop sequences [its DNA-C, -U1, and -S have slightly shorter (9 vs. 11 nts) stems than the other DNAs] but also in the sequences 5′ of the stem loop (Fig. 1); for instance, DNA-C and U1 of Mor23 lack the AatII site (GACGTC), which is not only present in all the DNAs of Mor5 and FBNSV-Eth but also conserved and unique in all bona fide genomic DNAs of FBNYV, MDV and SCSV, except for two integral DNAs of SCSV [17, 43]. For each DNA, the position of the potential promoter (TATA box) sequence TATATAA preceding the ORFs is indicated in Table 2. Potential polyadenylation signal sequences (AATAAA), which are required for transcription termination [37], are found 3′ of the major ORF in all DNAs.
Footnotes
The online version of the original article can be found under doi:10.1007/s00705-009-0548-9.


