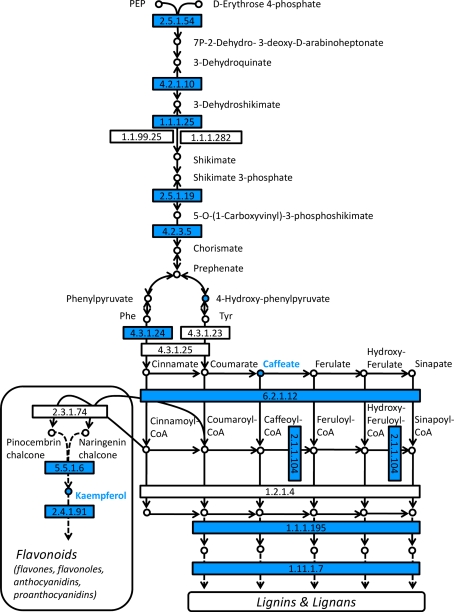
Fig. 5.
Analysis of pathways related to phenolics metabolism. Map displays selected steps from KEGG pathways pop00400 ‘Phenylalanine, tyrosin and tryptophan biosynthesis’, pop00940 ‘Phenylpropanoid biosynthesis’, pop00941 ‘Flavonoid biosynthesis’ and pop00944 ‘Flavone and flavonol biosynthesis’. Colors indicate significant expression, respective metabolite content ratios between P. x canescens PcISPS-RNAi line RA1 and wild type, blue indicates higher relative levels in wild type. Enzymes are given as EC numbers: 2.5.1.54, 3-deoxy-7-phosphoheptulonate synthase; 4.2.1.10, 3-dehydroquinate dehydratase; 1.1.1.25, shikimate dehydrogenase; 2.5.1.19, 3-phosphoshikimate 1-carboxyvinyltransferase; 4.2.3.5, chorismate synthase; 4.3.1.24, phenylalanine ammonia-lyase; 6.2.1.12, 4-coumarate:CoA ligase; 2.1.1.104, caffeoyl-CoA O-methyltransferase; 1.1.1.195, cinnamyl-alcohol dehydrogenase; 1.11.1.7, peroxidase; 5.5.1.6, chalcone isomerase; 2.4.1.91, flavonol 3-O-glucosyltransferase