Fig. 1.
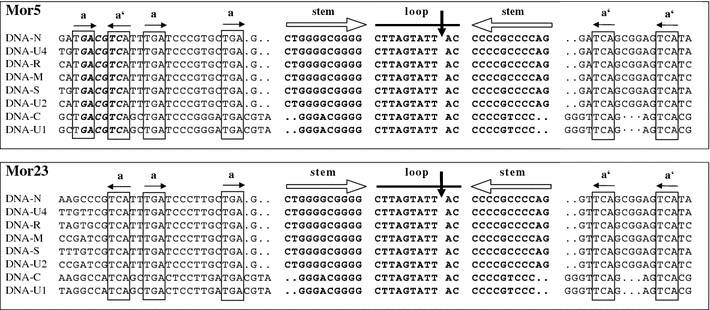
Alignment of the replication origin sequences of the eight DNAs of the isolates Mor5 and Mor23 illustrating the extent of sequence conservation in the stem-loop common region (CR-SL). Inverted repeat sequences (open horizontal arrows) potentially forming a stem-loop structure are in bold. A vertical arrow indicates the ‘origin of replication’, which is the position of cleavage by the master Rep protein [44]. Conserved iteron-like sequences (a) and their respective inversions (a′) possibly acting as recognition or binding sites for the master Rep protein are boxed and indicated by horizontal solid arrows. Dots indicate gaps included to maximise alignments. The AatII site (GACGTC) conserved in many nanoviruses [17] but absent from the Mor23 sequences is shown in bold italics in the Mor5 sequences
