Abstract
A long-term goal of mesenchymal progenitor cell (MPC) research is to identify cell-surface markers to facilitate MPC isolation. One reported MPC feature in humans and other species is lack of CD14 (lipopolysaccharide receptor) expression. The aim of this study was to evaluate CD14 as an MPC sorting marker. Our hypothesis was that cells negatively selected by CD14 expression would enrich MPC colony formation compared with unsorted and CD14-positive fractions. After validation of reagents, bone marrow aspirate was obtained from 12 horses. Fresh and cultured cells were analyzed by flow cytometry and reverse transcription and quantitative polymerase chain reaction to assess dynamic changes in phenotype. In fresh samples, cells did not consistently express protein markers used for lineage classification. Short-term (2-day) culture allowed distinction between hematopoietic and nonhematopoietic populations. Magnetic activated cell sorting was performed on cells from 6 horses to separate adherent CD14+ from CD14− cells. MPC colony formation was assessed at 7 days. Cells positively selected for CD14 expression were significantly more likely to form MPC colonies than both unsorted and negatively selected cells (P ≤ 0.005). MPCs from all fractions maintained low levels of CD14 expression long term, and upregulated CD14 gene and protein expression when stimulated with lipopolysaccharide. The equine CD14 molecule was trypsin-labile, offering a plausible explanation for the discrepancy with MPC phenotypes reported in other species. By definition, MPCs are considered nonhematopoietic because they lack expression of molecules such as CD14. Our results challenge this assumption, as equine MPCs appear to represent a descendant of a CD14-positive cell.
Introduction
Mesenchymal progenitor cells (MPCs) are found in bone marrow and other tissues, and can be defined using a number of criteria [1]. MPCs are adherent to tissue culture plastic and can differentiate to osteoblasts, adipocytes, and chondroblasts in vitro. Besides these features, human MPCs are defined by cell-surface expression of the cluster of differentiation (CD) molecules CD105, CD73, and CD90 and the lack of expression of CD45, CD34, CD14 or CD11b, CD79alpha or CD19, and HLA-DR in culture expanded cells [1]. Classification of MPCs using the CD cell surface phenotype has been utilized as supporting evidence of a unique cell population that can be distinguished from hematopoietic and other cell lineages.
One of the defining features of MPCs in humans and other species is the lack of expression of the cell-surface marker CD14, also known as the lipopolysaccharide receptor (LPS-R) [1]. Nevertheless, expression of CD14 cell surface molecule in MPCs and cells with similar differentiation properties is controversial. In 2003, Kuwana et al. described selection of CD14-positive cells from human peripheral blood that could then be differentiated in vitro into numerous mesenchymal tissues, including fat, bone, skeletal muscle, and cartilage [2], with a later study demonstrating differentiation into cardiomyocytes [3]. This group referred to the CD14-positive population of interest as monocyte-derived mesenchymal progenitors. Other groups have named peripheral blood cells with similar phenotype and differentiation capacity programmable cells of monocytic origin [4]. Pufe et al. demonstrated that these cells were able to form collagen type II producing chondrocytes in vitro. Using an antibody against the myeloid marker Mac-1 (also known as CD11b/CD18), Sera et al. demonstrated that murine adipocytes could be derived in vivo from hematopoietic cells of monocyte/macrophage lineage [5]. These studies and others challenge the assumption that cells capable of in vitro differentiation into mesenchymal tissues must lack expression of CD14.
The CD14 protein epitope is an important component of the innate immune system for detection of LPS. The LPS-R associates in a complex (53–55 kDa) with an adaptor protein known as myeloid differentiation protein-2 (MD-2) and the toll-like receptor 4 signaling proteins. The LPS-R has a molecular weight of 40 kDa when separated from the complex. Only certain lineages of hematopoietic cells (eg, monocytes, macrophages, dendritic cells, activated B lymphocytes, and, to a lesser extent, neutrophils) are known to express the CD14 cell surface molecule. Therefore, CD14 would be a candidate cell-surface marker to differentiate between adherent CD14-positive hematopoietic cells (primarily myeloid) and MPCs, as the latter should be negative for CD14 expression according to the proposed criteria used to define human MPCs [1].
Characterization studies of established human MPC cultures using differentiation assays, gene expression analysis, and cell surface protein markers have been performed for over a decade [6]. Most studies evaluate MPC cell surface protein markers and gene expression after long-term (several weeks) population expansion in culture to obtain sufficient cell numbers for analysis [7–10]. However, there are conflicting reports in MPC marker protein expression patterns when comparing phenotypes of freshly sorted MPCs to culture-expanded MPCs [11,12]. For example, putative MPCs from cord blood have been isolated with the initial cell phenotype of CD45+,CD105+, CD14+, CD49a+, CD49f+, CD44+, and CD34−. Later, the culture passaged cells were CD45−, CD14−, CD34−, and weakly CD105+ [13]. These studies, including previous work in our laboratory, suggest that the phenotype of MPCs is dynamic during isolation and culture processes [14].
If antibodies against specific cell surface molecules are to be used in bone marrow cell sorting assays, several important preliminary steps are necessary. The candidate antibody must first be validated for reactivity and specificity against the protein of interest. In addition, one must gain an understanding of the dynamic expression characteristics of the candidate cell surface molecule on cultured bone marrow cells. Understanding the protein expression pattern is needed to determine the time when cell sorting would be most efficient (eg, the candidate protein is differentially expressed between hematopoietic and nonhematopoietic cell populations). Since bone marrow cells are normally cultured for several weeks before MPC phenotyping, one of our goals was to evaluate freshly isolated bone marrow cells and bone marrow cells early in culture to study changes in protein expression that may have an impact on the efficiency and accuracy of cell sorting.
In the original characterization article by Pittenger et al., CD14 expression was reported as negative in human MPCs [6]. In that study, culture-expanded cells were harvested using either trypsin, a serine protease, or ethylenediaminetetraacetic acid (EDTA), with no reported difference in cell surface protein detection for any marker between cell dissociation solutions. Most subsequent MPC characterization studies have used the protocol described by Pittenger, including trypsinization, for cell harvest before flow cytometric analysis. Trypsin cleaves peptide chains at the carboxyl side of lysine or arginine. It is commonly used to disrupt adherent cells to permit cell passage or harvest in tissue culture applications. Trypsin has previously been shown to cleave a number of cell-membrane molecules, including CD14 [15]. Therefore, we wanted to evaluate the effect of trypsinization on detection of CD14 protein in equine MPCs.
In this study, our aim was to evaluate CD14 expression patterns in equine bone marrow-derived MPCs for use as a candidate marker for MPC enrichment. Our hypothesis was that bone marrow cells negatively selected for CD14 expression would enrich MPC colony formation compared with unsorted and CD14 positively selected cell fractions. This study contributes important information to the long-term goal of increased purity and decreased time for isolation of the putative MPC population.
Materials and Methods
Study design
The design of the experiment was divided into 3 phases. During Phase 1 the CD14 antibody was validated for protein reactivity and specificity in peripheral blood using flow cytometry and immunoprecipitation. Phase 2 of the experiment evaluated CD14 expression in bone marrow cells in freshly isolated and cultured cells overtime using flow cytometry and reverse transcription and quantitative polymerase chain reaction (qRT-PCR). In Phase 3, adherent bone marrow cells that had been cultured for 2 days were sorted based on their expression of CD14. Evaluation of colony formation, response to stimulation with LPS, and sensitivity of the CD14 epitope to trypsinization were subsequently compared between CD14 positively or negatively selected and unsorted fractions. An overview of the experiments detailing procedures, timelines, and animal numbers is presented in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertonline.com/scd). All procedures were performed in compliance with institutional guidelines for research on animals.
Phase 1
CD14 antibody validation
The mouse antiequine CD14 antibody (clone 105; B. Wagner, Cornell University) was tested for reactivity and specificity with the equine CD14 cell surface molecule. Whole blood (30 mL) was collected from 2 horses for antibody validation of reactivity with equine cells. Blood samples were drawn into heparin to a final concentration of 33 units/mL. Half of each blood sample was incubated with carbonyl iron powder (C3518; Sigma-Aldrich, Saint Louis, MO) using 0.08 g of powder/mL of whole blood for 30 min at 4°C to remove the majority of neutrophils. The neutrophil depleted and whole blood fractions were subsequently processed using density gradient centrifugation to remove the majority of red blood cells as previously described [16]. Isolated peripheral blood cells were analyzed to evaluate reactivity of the antibody with neutrophil, lymphocyte, and monocyte populations using flow cytometry. For CD14 specificity analysis, whole cell lysates were prepared from fresh peripheral blood leukocytes, and from red blood cells with platelets (negative control) from 2 additional horses. To determine if the equine CD14 antibody bound a protein of the expected size (40 kDa), immunoprecipitation and Western blot analysis was performed. A portion of the white blood cell (WBC) lysate was also immunoprecipitated using an antibody known to recognize human CD14 (mouse anti-human CD14, clone biG 10, catalog # 021-1c.2; Biometec, Griefswald, Germany) and previously validated for cross-reactivity and specificity with equine cells [17,18] was used as a positive control. The anticipated CD14 protein size was based on previous literature, size similarity to other species, and predicted equine sequences. A 15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis gel was used to resolve the immunoprecipitated products. After protein transfer, the polyvinylidene fluoride membrane was probed with the biG 10 antibody in Western blot analysis.
Phase 2
Bone marrow aspirate collection and cell isolation
Bone marrow aspirate was withdrawn from the sternabrae of 12 horses (6 males and 6 females, age range 6 months–20 years) under standing sedation with xylazine hydrochloride (0.55 mg/kg IV) and local anesthesia using 2% lidocaine hydrochloride (10 mL/site). At least 2 separate sternabrae were sampled with no more than 30 mL withdrawn from each site to minimize dilution of the aspirate with peripheral blood. Samples were collected in heparin to a final concentration of 33 units/mL.
Bone marrow aspirate (60 mL) from each horse was diluted to 180 mL total volume using phosphate buffered saline plus 0.5% bovine serum albumin (PBS + BSA). The WBC fraction of the sample was enriched and the majority of red blood cells removed by layering each 30 mL aliquot of dilute sample on Ficoll-Paque Plus (Amersham Biosciences, Piscataway, NJ) for density gradient centrifugation, as described for antibody validation. Samples were resuspended in 50 mL MPC culture media [Dulbecco's modified Eagle's medium, glucose at 1,000 mg/L, containing 2 mM L-glutamine, penicillin (100 units/mL), streptomycin (100 units/mL), basic fibroblastic growth factor (bFGF, 1 ng/mL), and 10% fetal bovine serum] before cell counting using a hemocytometer. Approximately 2 to 9 × 108 bone marrow mononuclear cells (BMMNC) were obtained per sample (3–15 × 106 BMMNC/mL of bone marrow aspirate).
MPC expansion in culture
BMMNCs were plated onto 10-cm-diameter tissue culture plates at a density of ∼300,000 cells/cm2 (20 × 106 cells/plate). Cells were cultured in MPC culture media at 37°C in a 5% CO2, 95% air atmosphere at 5% humidity. One-half of the medium (5 mL) from all plates was removed at 24 h of culture and replaced with a fresh medium. Subsequently, media were exchanged every 72–96 h. At subconfluence of 70%–90%, cells were passaged 1:3 using Accumax® cell dissociation solution (Innovative Cell Technologies, Inc., San Diego, CA) at a concentration of 1 mL/15 cm2, and replated at a density of 6,000–10,000 cells/cm2. Cells were incubated with Accumax solution for 5 min at 37°C. Accumax solution was used to avoid potential damage to cell surface proteins and to prevent cellular clumping. Samples were collected at 2 h and on days 2, 5, 7, 14, 21, and 30 of culture for flow cytometric and quantitative gene expression analysis. Cells were analyzed at these time points to evaluate the changes in cell surface protein expression over time.
Flow cytometric analysis of cell surface markers
For the experiments herein, aliquots of cells (106) were labeled for CD14 and other cell surface molecules (CD44, CD90, CD11a/CD18, and CD172a) known to vary in expression on bone marrow cells over time in culture using monoclonal antibodies from a panel previously validated for the horse [14]. A portion (∼107 cells) of the freshly isolated bone marrow aspirate samples from all 12 horses were analyzed using flow cytometry (n = 12). The remaining portion of bone marrow cells were then placed into tissue culture. Samples from 5 of the horses were collected after 2 h of culture. Analysis of both the adherent and nonadherent cell fractions were compared with results from analysis of freshly isolated cells (n = 12). The remaining (n = 7) bone marrow samples continued in culture and adherent cells were harvested on 2, 7, 14, 21, and 30 days.
Cells were treated with a 20-min blocking step using 10% normal goat serum in fluorescence-activated cell sorting (FACS)-Buffer (phosphate buffered saline containing 2.5% fetal bovine serum). The cells were washed with FACS-Buffer, resuspended in unconjugated primary monoclonal antibody, and incubated for 45 min at 4°C. Cells were then washed and incubated with a second fluorescent-conjugated goat anti-mouse IgG or IgM antibody [fluorescein isothiocyanate (FITC)-conjugated AffiniPure goat anti-mouse IgG (H+L) or IgM μ Chain Specific; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA] for an additional 45 min at 4°C. Cells not treated with antibody, and cells incubated with mouse antiparvovirus antibody, and FITC secondary antibodies were used as negative controls. Cells were washed, resuspended in FACS-Buffer, and analyzed on a FACSCalibur (Becton Dickinson Immunocytometry Systems, San Jose, CA) flow cytometer equipped with a 488 nm argon laser, and BD Cell Quest™ analysis software (BD Biosciences, San Jose, CA). The settings for the flow cytometric analyses were calibrated to determine <2% positive cells for the control antibodies. Percentage positive and mean fluorescence intensity data were collected on 105 cells for each sample regardless of size and granularity (nongated) to prevent bias in gating. Dot plot settings for uncultured bone marrow cells and cells cultured 2 h were based on a linear scale.
For culture-expanded cells, flow cytometric analysis was performed on days 2, 7, 14, 21, and 30 after isolation. Supernatant was removed and adherent cells were lifted from the plate using Accumax. Cultured cells were processed and analyzed by flow cytometry as described above. The only variation was that dot plot settings were adjusted to a logarithmic scale in the cultured cells to include large, granular cells. Flow cytometric analysis of cell surface molecule expression was performed in the gate determined to contain dividing cells based on the results from the propidium iodide DNA staining assay as previously described [14].
RNA extraction and one-step qRT-PCR
Gene expression analysis was performed to confirm negative protein results and account for kinetic changes in transcription and translation. At the same culture time points when cells were analyzed by flow cytometry, RNA was extracted from ∼1 to 3 × 106 cells of the corresponding samples using either Trizol® (Life Technologies/Invitrogen, Carlsbad, CA) or the 5 Prime Perfect Pure RNA® extraction kit (5 Prime, Inc., Gaithersburg, MD) according to the manufacturers' directions. Cells from the fresh and 2-h samples were not analyzed using qRT-PCR because of the heterogeneous cell populations present. RNA quantity and quality were determined using a Nanodrop® spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE), and observation of 18 and 28S bands on 0.8% agarose gels.
RNA samples were diluted to a concentration of 20 ng/well; 2 replicate wells were used for quantitative gene expression assays. Total RNA was reverse transcribed and amplified using the 1-step RT-PCR technique and the ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA). The primers and dual-labeled fluorescent probe [6-FAM as the 5′ label (reporter dye) and TAMRA as the 3′ label (quenching dye)] were designed using Primer Express Software Version 2.0b8a (Applied Biosystems). The CD14 probe and primers were designed using equine-specific sequences published in GenBank, and the sequence obtained in our laboratory. A portion of the CD14 gene was cloned and gene sequence agreed with previously reported data (AF200416). The 18S ribosomal subunit was used to normalize gene expression, as ribosomal RNA has previously been shown to be a reliable housekeeping gene in quantitative gene expression analyses with less individual and tissue variability than other housekeeping genes such as glyceraldehyde 3-phophate dehydrogenase or β-actin [19,20]. The 2−ΔΔCt method was used for relative gene expression analysis. A sample from unsorted bone marrow cells cultured 14 days was used as the calibrator sample since this was the earliest time point in culture when MPCs were the predominant cell type present. The 18S gene had the following primers and probe:
Forward, 5′-CGGCTTTGGTGACTCTAGATAACC-3′
Probe FAM, (5′)-TCGAACGTCTGCCCTATCAACTTTCGAT-TAMRA (3′)
Reverse, 5′-CCATGGTAGGCACAGCGACTA-3′ (GenBank accession no. AJ311673).
CD14 had the following primers and probe:
Forward, 5′-TACGTGCGCTCGGGTACTC-3′
Probe FAM, (5′)-CGCCTCAAGGAACTGACGCTGGA-TAMRA (3′)
Reverse, 5′-CATCGTGCCGGTTACCTCTAG-3′ (GenBank accession no. AF200416).
Quantitative RT-PCR was performed to provide supporting evidence that CD14 expression levels were consistent with cell surface CD14 protein expression at the different culture time points during Phase 2 of the study, and later during Phase 3 to evaluate gene expression of the sorted and unsorted cell fractions.
Phase 3
Magnetic activated cell sorting
Bone marrow aspirate was collected from 6 horses (3 males and 3 females, age range 3–10 years) and processed using density gradient centrifugation. After mononuclear cell isolation, bone marrow cells were cultured for 48 h. At 48 h, the plates were washed twice with 5 mL of media to remove nearly all nonadherent cells before harvest using Accumax cell dissociation solution. The MidiMAC system from Miltenyi Biotec, Inc. (Auburn, CA) was used for subsequent MACS of bone marrow cells. Adherent cells were consolidated and resuspended in 500 μL of chilled and degassed MACS buffer (0.5% bovine serum albumin in phosphate buffered saline with 2 mmol EDTA to prevent clumping). To remove any cell clumps before antibody labeling, samples were gravity filtered over MACS® preseparation filters (Miltenyi Biotec, Inc.). After a cell count, approximately one-third of the sample was removed as the unsorted fraction for flow cytometric analysis, culture, and RNA extraction. The remaining two-thirds of the sample was pelleted and resuspended in MACS buffer and mouse antiequine CD14 antibody (90 μL buffer + 10 μL of antibody/107 cells), as this is the dilution used for flow cytometry. The primary antibody was incubated with cells for 10 min at 4°C. The labeled samples were washed twice with 2 mL of MACS buffer/107 cells followed by 5-min centrifugations at 500 g. Next, cell pellets were resuspended in 80 μL MACS buffer + 20 μL of rat anti-mouse IgG MACS microbeads/107 cells. Samples were incubated for 15 min at 4°C and then were washed with 2 mL of MACS buffer/107 cells followed by 5-min centrifugation at 500× g. The LS column, fit with a new preseparation filter, was placed in the magnetic field and primed using 3 mL of chilled, degassed MACS buffer. A 20 gauge needle was affixed to the end of the column to slow flow of liquid through the column and prevent washout of positive cells. The pellet was resuspended in 500 μL of MACS buffer and placed over the primed preseparation filter and LS column. The CD14-negative fraction was collected as the flow-through portion of the sample. The column was allowed to drain until the flow-through stopped dripping between all steps. Three washings with 3 mL of MACS buffer each were used to rinse additional negative cells from the column. The CD14-positive fraction of the sample was collected after removal of the column from the magnetic field. The column was loaded with 5 mL of MACS buffer and a plunger was used to force positive cells from the column into a collection tube. Positive and negative fractions were counted and the samples divided for flow cytometric analysis, culture, and RNA extraction.
Assessment of sort purity
To assess the MACS unsorted fraction, ∼8 × 106 cells were collected for flow cytometry analysis using a previously validated panel of monoclonal antibodies and the CD14 antibody as previously described [14]. The positive and negative sorted fractions had 2 × 106 cells allocated for flow cytometry analysis. Half of the cells (106) of each sorted fraction remained unlabeled with fluorescent secondary; the other 106 cells were labeled with goat anti-mouse IgG FITC secondary antibody.
Colony formation assay
For all 3 fractions (CD14 positive, CD14 negative, and unsorted) 1.15 × 106 cells were collected for quantitative analysis of colony formation after 1 week of culture. A plating density of 20,000/cm2 in 6-well plates was used. Routine culture commenced as described above. On day 7 of culture, the numbers of colonies present in each fraction were counted. A colony was defined as a cluster of 50 or more fibroblastic cells. Colony numbers were compared between the unsorted and CD14 positively and negatively selected fractions.
qRT-PCR during Phase 3
CD14 expression data were compared over time between unsorted, positive, and negative selected cells. Day 14 of culture was the first time point in which sufficient MPC numbers were available in all 3 fractions for gene analysis (negative fraction samples often had too few cells at the 2 and 7 day time periods). Limited numbers of earlier time point samples were included when available to verify sorting efficiency. In a later experiment, CD14 expression was also compared between sorted MPC fractions harvested with trypsin or Accumax solutions to confirm that CD14 gene expression in these samples was consistent with other MPC samples cultured for a similar time period.
Analysis of CD14 expression in cultured MPCs in response to LPS stimulation
Bone marrow cells from 1 horse were isolated, cultured, and MACS sorted as described above. At 21 days, a portion of the cells from the CD14-positive and the unsorted fractions were incubated overnight with LPS (026:B6, L2762; Sigma-Aldrich) at varying dosages (0, 1, 5, or 10 ng/mL media). Samples of control and LPS-treated cells were collected for flow cytometric analysis of CD14 protein expression and RNA extraction for qRT-PCR analysis of CD14 expression.
Analysis of cell surface marker expression in response to trypsinization
Bone marrow cells from 4 horses were isolated, cultured, and MACS sorted as described above. At 30 days of culture, ∼107 cells from each fraction (CD14 positive, CD14 negative, or unsorted) were harvested using Accumax cell dissociation solution. An additional 107 cells from each fraction were harvested after 5 min of incubation at 37°C using 0.25% trypsin in Hanks balanced salt solution with divalent cations. Cells (2 × 106) from each sample were used for RNA extraction and subsequent qRT-PCR. The remaining 8 × 106 cells were used for flow cytometric analysis with the same panel of antibody markers as previously described to determine if trypsinization would have an effect on their mean fluorescence intensity.
Statistical analysis
Phase 1 of the study was purely descriptive, and no statistical analysis was performed.
Phase 2 CD14 gene expression data were categorized into 4 groups by culture duration: 1 = <1 week; 2 = 1 week; 3 = 2 weeks; 4 = 3 weeks or more. Groups were compared using a 1-way ANOVA with a Tukey all-pairwise comparisons post hoc test. A P value of <0.05 was considered significant.
Phase 3 had statistical analyses performed on the data from several assays. Colony formation data (colonies/106 cells) were categorized into 3 groups by cell sorted fraction: 1 = unsorted; 2 = CD14 positive; 3 = CD14 negative. Groups were compared using a 1-way ANOVA with Tukey All-Pairwise Comparisons post hoc test. The CD14 gene expression data of MACS sorted versus unsorted cells were categorized by culture duration: 1 = <1 week; 2 = 1 week; 3 = 2 weeks; 4 = 3 weeks; 5 = 4 or more weeks. Groups were compared using a 1-way ANOVA with Tukey All-Pairwise Comparisons post hoc test. LPS cell stimulation data were categorized by cell fraction (1 = unsorted; 2 = CD14 positive) and LPS dose (1 = 0 ng/mL; 2 = 1, 5, or 10 ng/mL LPS). Groups were compared using a 1-way ANOVA with Tukey All-Pairwise Comparisons post hoc test. Flow cytometric data of the percentage of positive cells in each cell fraction (1 = unsorted; 2 = CD14 positive; 3 = CD14 negative) were compared between cells lifted with 1 = Accumax and 2 = 0.25% trypsin for the panel of antibody markers. Groups were compared using a 1-way ANOVA with Tukey All-Pairwise Comparisons post hoc test. A P value of <0.05 was considered significant.
Results
Phase 1
Validation of antibody binding to the CD14 molecule on equine peripheral blood cells
The equine CD14 antibody detected positive equine blood cells using flow cytometric analysis (Fig. 1). As expected, there was low mean fluorescence intensity detected for CD14 in the neutrophil population (Fig. 1, R1) and high mean fluorescence intensity in the monocyte population (Fig. 1, R3). A small population of cells in the lymphocyte gate (Fig. 1, R2) had low mean fluorescence intensity, which likely represent active B lymphocytes; double labeling analysis would confirm that these cells are B lymphocytes.
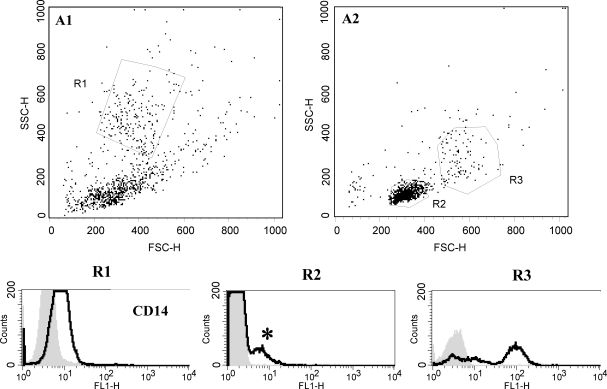
FIG. 1.
Flow cytometric analysis of CD14 cell surface molecule expression of epitope reactivity in freshly isolated peripheral blood leukocytes. Dot plot distribution of uncultured peripheral blood cells isolated using (A1) gradient density centrifugation or (A2) carbonyl iron incubation followed by gradient density centrifugation. The R1 gate corresponds to the size and granularity of neutrophils, R2 lymphocytes, and R3 monocytes. (R1–R3) Histogram analysis of mean fluorescence intensity of CD14 cell surface molecule expression in the gated areas. The shaded curves represent negative isotype control staining in each cell population. Open lines represent the mean fluorescence intensity for CD14. Neutrophils (R1) have low mean fluorescence intensity for CD14 expression, whereas monocytes (R3) have high mean fluorescence intensity. The small population of lymphocytes (R2) with low mean fluorescence intensity likely represents the activated B lymphocyte population (*).
Validation of CD14 specificity in equine peripheral blood cells
Western blot analysis alone did not clearly determine specificity of the mouse antiequine CD14 antibody (B. Wagner, Cornell University, clone 105) reaction with the CD14 epitope. No distinct band was detected on multiple attempts to analyze CD14 antibody binding (data not shown) using equine peripheral blood lysates. It was proposed that this antibody might not recognize the denatured protein. The biG10 antibody has been extensively used for CD14 immunoprecipitation in human cells. Immunoprecipitation using the mouse antiequine CD14 antibody (clone 105) followed by Western blot analysis with the mouse anti-human biG10 antibody confirmed that the CD14 antibody (clone 105) reacted with an equine protein of ∼40 kDa (Fig. 2). The protein size was comparable to the immunoprecipitated product when biG 10 antibody was used on the equine WBC lysate.
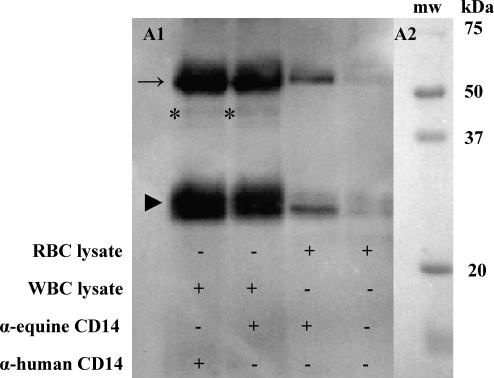
FIG. 2.
Immunoprecipitation and Western blot analysis of specificity for CD14 antibody to the equine molecule expressed in peripheral blood cells. (A1) Equine peripheral blood Western blot probed with mouse anti-human CD14 biG10 antibody after immunoprecipitation using either anti-(α)-equine CD14 antibody or biG10 antibody with equine blood cells. WBC lysates immunoprecipitated with both CD14 antibodies retained protein bands, of ∼40 kDa (*). Arrow indicates the heavy chain of IgG; arrowhead indicates the light chain of IgG. (A2) Molecular size standards. WBC, white blood cell. RBC, red blood cell.
Phase 2
Flow cytometric analysis of cell surface protein expression in bone marrow cells freshly isolated and cultured for 2 h
There was a slight shift in dot plot population distributions between freshly isolated cells and cultured nonadherent cells at 2 h (eg, there were slightly higher percentages of cells in Region 2 in nonadherent cells compared with freshly isolated cells) (Fig. 3A, B). In contrast, at 2 h, cultured adherent bone marrow cells had a unique dot plot distribution (Fig. 3C) enriched in Region 3 compared with freshly isolated and nonadherent cells.
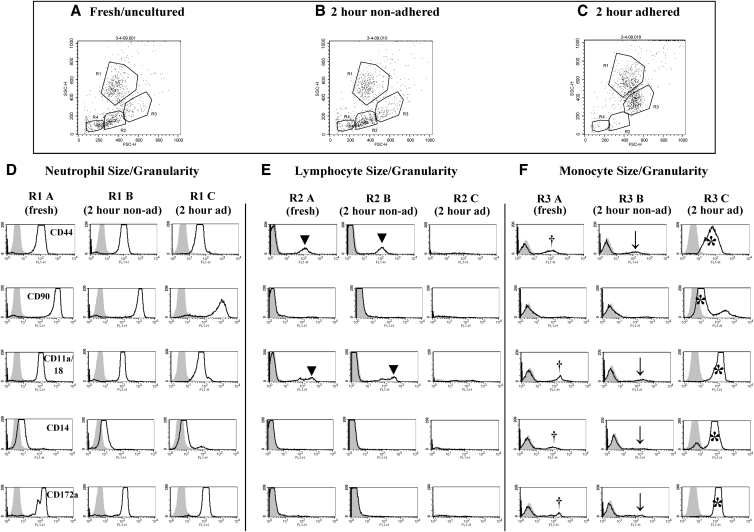
FIG. 3.
Flow cytometric analysis of cell surface protein expression in freshly isolated bone marrow cells compared with cells cultured for 2 h. (A–C) Dot plot distribution of bone marrow cells isolated using gradient density centrifugation (A) fresh/uncultured, (B) nonadherent cultured, or (C) adherent cultured cell fractions at 2 h of culture. (D–F) Histogram analysis of mean fluorescence intensity of cell surface molecule expression in the gated areas (Regions 1, 2, and 3, respectively). (D) R1 designates cells of the size/granularity equivalent to neutrophils; (E) R2 designates cells of the size/granularity equivalent to lymphocytes; (F) R3 designates cells of the size/granularity equivalent to monocytes. The shaded curves represent negative isotype control staining; open lines represent the labeling for the cell surface markers indicated on the left-hand side. A small fraction (about 15%) of the lymphocyte-like bone marrow cells from both freshly isolated and 2-h nonadherent cultures was positive for CD44 and CD11a/CD18 expression (▾). In the monocyte-like bone marrow cells, about 15% of freshly isolated (†) or nonadherent cells (↓) had detectable expression of some molecules. In contrast, after 2 h of culture, the monocyte-like adherent bone marrow cells (*) had a distinct phenotype.
There were no notable differences in cell surface molecule expression for all markers in bone marrow cells equivalent in size and granularity to neutrophils both before and after culture (Fig. 3D). Neutrophil-like cells in all fractions displayed a phenotype of CD44hi, CD90hi, CD11a/CD18hi, CD14low, and CD172ahi. This phenotype was consistent with the phenotype of equine peripheral blood neutrophils. In lymphocyte-like bone marrow cells, there were differences in histogram distribution for all CD markers analyzed between freshly isolated or 2-h nonadherent cells and 2-h adherent cells since there were very few lymphocyte-like cells in the adherent cell fraction (Fig. 3E). A small fraction (∼15%) of lymphocyte-sized bone marrow cells from both freshly isolated and 2-h nonadherent cultures expressed CD44 and CD11a/CD18 (Fig. 3E). An absence of CD90, CD14, and CD172a protein expression was consistent with the expected phenotype of peripheral blood lymphocytes. In 2-h adherent monocyte-like bone marrow cells, there was a marked difference in cell surface protein expression for all markers when compared with freshly isolated and 2-h nonadherent cells (Fig. 3F). Similar to lymphocyte-like bone marrow cells, only about 15% of freshly isolated or nonadherent monocyte-like cells were positive for CD44, CD11a/CD18, CD14, and CD172a molecules. In contrast, after 2 h of culture, the adherent monocyte-like bone marrow cells had a distinct phenotype of CD44hi, CD90low/mod, CD11a/CD18hi, CD14hi, and CD172ahi, consistent with the phenotype of equine peripheral blood monocytes.
Flow cytometric analysis of cell surface marker expression in cultured bone marrow cells
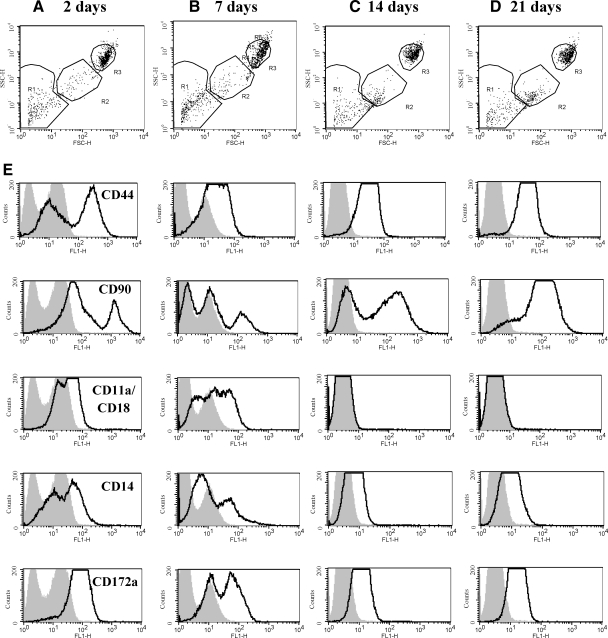
After 2 days of culture, adherent mononuclear cells displayed an antibody labeling pattern of CD44hi, CD90mod/hi, CD11a/CD18mod, CD14mod, and CD172ahi (Fig. 4). In Gate 3, there was a heterogeneous cell population indicated by multiple peaks in CD44, CD90, CD11a/CD18, and CD14 mean fluorescence intensity. On day 7, CD44 expression slightly decreased in mean fluorescence intensity, but remained positive and became more homogenous. CD90 expression was negative in 2 of the 3 cell populations present, but high mean fluorescence intensity was noted in a third population. Meanwhile, CD11a/CD18, CD14, and CD172a cell surface protein expression decreased, with a shift in the populations to lower mean fluorescence intensity. By 14 days, adherent cells were CD44hi, CD90hi/variable, CD11a/CD18neg, CD14low, and CD172alow; these cells displayed a fibroblastic morphology characteristic of MPC (data not shown). Multiple cell populations (multiple peaks) were still noted in CD90 mean fluorescence intensity at 14 days, with a growing number of positive cells compared with 7 days. All other cell surface markers had homogeneous protein expression by 14 days. The pattern of molecule expression was retained at 21 days for all MPC samples, with CD90 becoming homogenous with high mean fluorescence intensity. Throughout time in culture (2, 7, 14, and 21 days), adherent bone marrow cells developed a homogeneous protein phenotype.
FIG. 4.
Flow cytometric analysis of cell surface molecule expression in cultured bone marrow cells from 2 through 21 days of culture. (A–D) Dot plot distribution of bone marrow cells cultured for (A) 2 days, (B) 7 days, (C) 14 days, or (D) 21 days. (E) Histogram analysis of mean fluorescence intensity of surface molecule expression in Region 3 (R3) cells. The shaded curves represent negative isotype control staining; open lines represent the labeling for the cell surface markers indicated in the left-hand side.
Gene expression kinetics of CD14 in cultured bone marrow cells over time
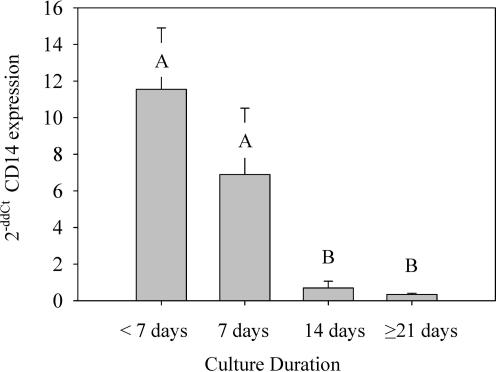
CD14 gene expression data were consistent with cell surface protein expression at all culture time points. CD14 expression was present in all samples but became significantly less (P ≤ 0.005) over time (Fig. 5). Samples cultured 1 week or less had significantly more CD14 expression than samples cultured for >1 week. CD14 expression remained stable at low levels after 14 or more days of culture, with <1-fold change compared with the calibrator sample.
FIG. 5.
Gene expression kinetics of CD14 during cell culture. A bone marrow sample cultured for 14 days was used as the calibrator sample for comparison using the 2−ΔΔCt method. Bars represent n = 6 ± SE, letters A and B denote significant differences between groups.
Phase 3
Magnetic activated cell sorting using the mouse anti-horse CD14 antibody
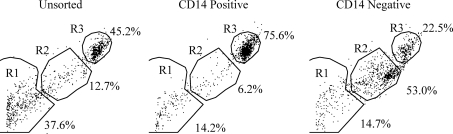
MACS of adherent bone marrow cells after 2 days of culture led to distinct flow cytometric dot plot distributions of cells within the CD14-positive and -negative fractions compared with unsorted conditions (Fig. 6). The CD14-positive fraction cells were concentrated in Region 3, whereas CD14-negative selection cells were concentrated in Region 2.
FIG. 6.
Flow cytometric dot plot analysis of equine bone marrow cells cultured for 2 days, after MACS sorting using a mouse antiequine CD14 antibody. The percentage of cells in each region are listed.
Quantification of colony formation after cell sorting using CD14 monoclonal antibody
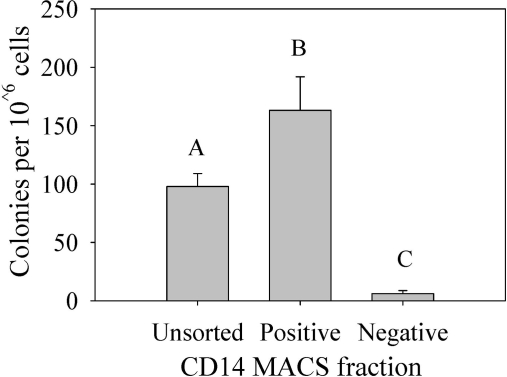
After a week of culture, a significant difference in MPC colony formation was noted between all 3 fractions (P ≤ 0.005) (Fig. 7). Cells positively selected for CD14 expression had significantly more MPC colonies than either the unsorted or negatively selected fractions, with nearly double the colony counts of unsorted cells and over 20 times the number of colonies formed in the CD14 negatively selected fractions.
FIG. 7.
Quantification of colony formation after cell sorting using CD14 monoclonal antibody. The number of MPC colonies formed per 106 cells in each fraction were compared between groups. Bars represent n = 6 ± SE, letters A, B, and C denote significant differences between groups. MPC, mesenchymal progenitor cell.
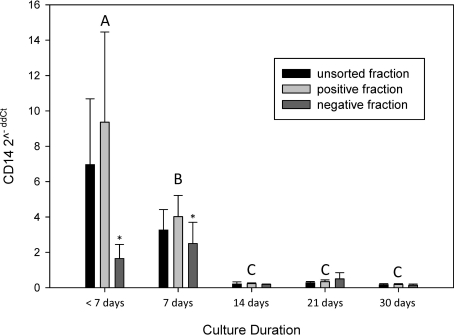
CD14 gene expression over time between sorted and unsorted cells
Sufficient cell numbers (≥50,000) in the negatively selected fraction before 14 days of culture for analysis were difficult to obtain (n = 3). Samples cultured 7 days or less had significantly more CD14 expression compared with established MPC cultures (P ≤ 0.005) (Fig. 8). Since CD14 expression of samples were being compared with an unsorted sample cultured 14 days, cells from all 3 fractions (unsorted, positive, and negative) cultured 7 days or less had significantly more CD14 expression than the calibrator sample (P ≤ 0.005). There were no significant differences between CD14 expression in any of the 3 fractions at 14 (P = 0.38), 21 (P = 0.69), or 30 (P = 0.94) days of culture when MPCs were the predominant cell type in the samples.
FIG. 8.
CD14 gene expression over time between CD14 sorted and unsorted MACS cell fractions on adherent bone marrow cells. A reduced number of samples (n = 3) were available in the negative fractions of samples cultured 7 days or less (*). An unsorted bone marrow sample cultured for 14 days was used as the calibrator sample for comparison using the 2−ΔΔCt method. Bars represent n = 6 ± SE, letters A, B, and C denote significant differences between time period groups.
Flow cytometric analysis of CD14 expression in cultured MPCs in response to LPS stimulation
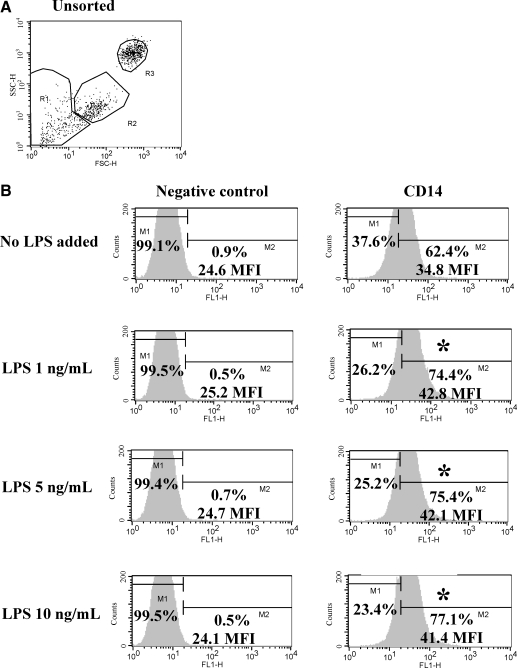
There were no detectable differences in flow cytometric CD14 mean fluorescence intensity between unsorted and CD14 positively selected MPCs after 21 days of culture, with or without overnight LPS stimulation (data not shown). All doses of LPS (1, 5, or 10 ng/mL media) caused a significant increase in both percentage of positive cells (P = 0.013) and mean fluorescence intensity (P = 0.018) compared with untreated control cells (Fig. 9). Equine MPCs responded to LPS stimulation with an upregulation of CD14 protein expression.
FIG. 9.
Flow cytometric analysis of CD14 cell surface molecule expression in unsorted bone marrow cells in response to LPS stimulation. (A) Dot plot distribution of unsorted bone marrow cells cultured for 21 days. (B) Histogram analysis of CD14 mean fluorescence intensity in Region 3 after overnight incubation of cells with LPS of differing concentrations (0, 1, 5, or 10 ng/mL media). The histograms represent cell surface molecule expression using either a negative isotype control antibody (left column) or the mouse antiequine CD14 antibody (right column). M1 represents the setting used for negative cell percentage calculations based on isotype control labeling. M2 represents the setting for positive cell percentage calculations. (*) A significant difference in percentage of positive cells and mean fluorescence intensity from the untreated sample. LPS, lipopolysaccharide; MFI, mean fluorescence intensity.
qRT-PCR analysis of CD14 gene expression in cultured MPCs in response to LPS stimulation
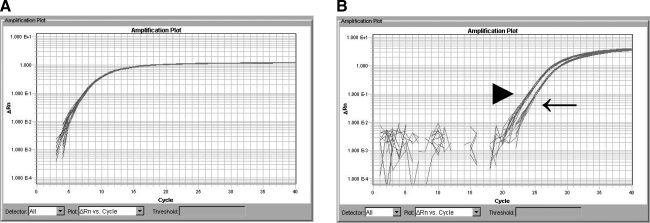
There were no detectable differences in CD14 expression between unsorted and positively selected MPCs after 21 days of culture using qRT-PCR. Both fractions had similar levels of CD14 expression in control samples, and displayed a similar increase in CD14 expression in response to LPS stimulation (Fig. 10). The 18S amplification plot demonstrated equal loading of RNA in all samples. Analysis of 2−ΔΔCt demonstrated a significant difference in CD14 expression between untreated and LPS stimulated cells in both unsorted and positively selected fractions (P ≤ 0.005). Cells treated with any dose of LPS (1, 5, or 10 ng/mL media) had a 2.24 (±0.12)-fold increase in CD14 expression over untreated cells. LPS stimulation promoted an increased CD14 expression in both unsorted and CD14-positive selected MPCs.
FIG. 10.
Gene expression kinetics of CD14 in unsorted or CD14-positive selected equine bone marrow cells with or without LPS stimulation. MPCs were cultured 21 days and treated with LPS at dosages of 0, 1, 5, or 10 ng/mL media. (A) 18S gene amplification plot (to verify equal RNA loading). (B) CD14 amplification plot of LPS stimulated and control bone marrow cells. Control MPCs treated with 0 ng/mL of LPS (←) had higher Ct values (lower levels of CD14 expression) than cells that were treated with LPS at any dose (▸).
Analysis of cell surface marker expression in response to trypsinization
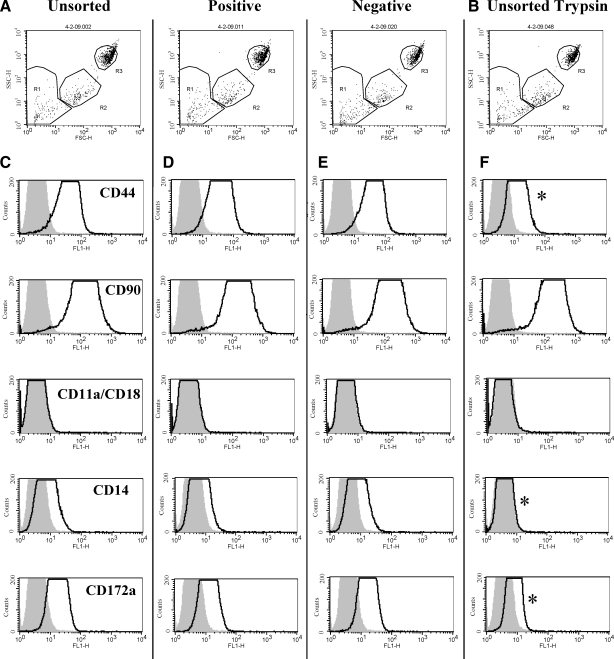
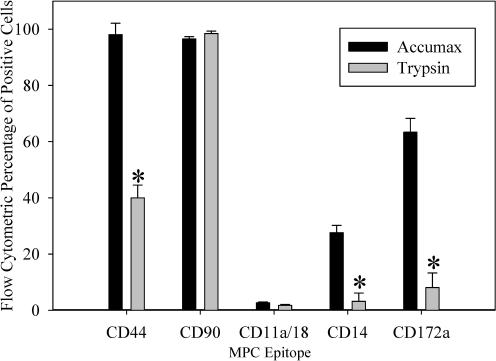
Flow cytometric dot plot analysis demonstrated no detectable difference in cell distributions between sorted and unsorted fractions cultured for 30 days (Fig. 11A, B). MPCs in all 3 fractions (unsorted, CD14 positive, or CD14 negatively selected) were CD44hi, CD90hi, CD11a/CD18neg, CD14low, and CD172alow, suggesting that the cells in each fraction are phenotypically similar at the 30 day culture time point when Accumax cell detachment solution is used for cell harvest (Fig. 11C–E). When 0.25% trypsin in Hanks balanced salt solution was used for cell harvest, there was a change in phenotype in all fractions to CD44low, CD90hi, CD11a/CD18neg, CD14neg, and CD172alow (Figs. 11F and 12). The mean fluorescence intensity for CD44 decreased in all samples, but protein expression was still present, and the percentage of CD44-positive cells dropped significantly (P ≤ 0.005) from 98.05% (±4.09) to 39.98% (±4.56). There appeared to be no effect on the mean fluorescence intensity for CD90 expression with trypsinization. The low expression level of CD11a/CD18 did not allow for any conclusions to be drawn on the effect of trypsinization. The low mean fluorescence intensity of CD14 expression detected in cells that were harvested using Accumax disappeared after trypsinization, suggesting that the equine CD14 epitope is trypsin-labile. The percentage of CD14-positive cells was reduced from 27.60% (±2.61) to 3.18% (±2.92) after incubation with trypsin, causing the trypsin-treated samples to appear negative for CD14 expression (P ≤ 0.005). Meanwhile, CD14 gene expression was present in comparable quantities between cells treated with trypsin or Accumax cell harvest solutions (data not shown). Finally, there was a significant decrease in CD172a percentages from 63.39% (±4.89) to 8.08% (±5.18) when trypsin was used for cell harvest (P ≤ 0.005). Harvesting cells for flow cytometric analysis with trypsin appears to damage several cell surface proteins, whereas others appear to be unaffected.
FIG. 11.
Flow cytometric analysis of cell surface molecule expression in sorted and unsorted bone marrow cells treated or not with trypsin. (A, B) Dot plot distribution of bone marrow cells cultured 30 days after separation at 2 days of culture into unsorted, positively, or negatively selected fractions using an antiequine CD14 antibody and MACS. Adherent cells were collected for analysis after 5 min of incubation at 37°C with either Accumax® cell detachment solution (A) or 0.25% trypsin in Hanks balanced salt solution (B). (C–F) Histogram analysis of mean fluorescence intensity of cell surface molecule expression in Region 3. Shaded curves represent negative isotype control staining; open lines represent labeling for the cell surface markers indicated on the left-hand side. When trypsin was used to collect the cells for analysis, a decrease in mean fluorescence intensity was noted for some antibodies (F, *).
FIG. 12.
Flow cytometric analysis of cell surface molecule expression in bone marrow cells cultured for 30 days and treated with either Accumax or 0.25% trypsin in Hanks balanced salt solution. The percentage of positive cells using the respective antibody was compared between solutions (n = 4 ± SE). There were significant reductions (P ≤ 0.005) in the percentage of positive cells for some molecules (*) when trypsin was used instead of Accumax for cell preparation. The 0.25% trypsin solution reduces the detection of some, but not all cell surface markers in flow cytometric analysis.
Discussion
In this study, a number of important findings were documented. We demonstrated differences in cell surface protein expression and population distributions during the analysis of freshly isolated bone marrow cells compared with cultured cells. The temporal analysis and controlled sample processing under the culture conditions presented herein support the results that clearly showed CD14 protein expression was present in equine bone marrow-derived putative MPCs throughout culture, unless the CD14 epitope was damaged by trypsinization during processing. We have also documented that bone marrow cells positively selected for CD14 expression were significantly more likely to form MPC colonies than both unsorted and negatively selected cells. Our results challenge the classification of putative equine MPCs as CD14 negative.
Early in Phase 2 of the study we sought to determine the optimal culture duration to perform subsequent cell sorting assays. Comparison of freshly isolated bone marrow cells with cells that were primarily adherent or nonadherent after 2 h of culture showed clear differences in expression of CD14 and other cell surface marker proteins. For example, the mean fluorescent intensity of cell surface marker expression and the percentages of positive cells were higher in the adherent monocyte-like population when compared with cells that were monocyte-sized from the freshly isolated or nonadherent fractions, suggesting that the short culture duration of 2 h was useful in separating bone marrow cells types at varying stages of differentiation. These results suggest that separation of freshly isolated bone marrow cells based on expression of certain cell surface proteins (eg, CD11a/CD18, CD44, or CD14) may lead to inaccurate classification of hematopoietic versus nonhematopoietic cells, and could lead to contamination with hematopoietic cells in putative MPC cultures. Caution should be exercised when evaluating results of studies that attempt to sort hematopoietic from nonhematopoietic cells using cell surface molecules characteristic of peripheral blood cells in uncultured bone marrow samples.
One of the defining features of MPCs in humans and other species is lack of expression of the CD14 epitope on their cell surface. However, our results clearly demonstrated that CD14 is expressed on the surface of putative equine MPCs. Mean fluorescence intensity and CD14 gene expression decrease significantly in established MPC populations compared with cells cultured for 7 days or less; however, after 14 days of culture, CD14 expression at both the protein and gene level was stable. This is in contrast to flow cytometric analysis of cell surface markers such as CD11a/CD18, which was negative in equine MPCs cultured 14 or more days. All RNA samples analyzed by qRT-PCR were able to reach a Ct value for CD14 expression, unlike CD11a and CD45 in a previous study [14], suggesting that CD14 expression is present, at low levels, in equine MPCs.
Contrary to our hypothesis, the MPC population was not enriched in the CD14-negative fraction of cultured equine bone marrow cells using MACS. Further, CD14 expression can be used as a marker to enrich for the MPC population when MACS is performed on equine bone marrow cells after 2 days of culture. Flow cytometry was a useful technique to assess the initial purity of sorted cells after MACS. Previous work from our laboratory [14] suggested that MPCs came from the region concentrated in the CD14-positive fraction. The percentage of positively selected cells located in this region was nearly double the percentage of unsorted cells, and 3–5 times more than the negatively selected cell fraction. Comparison of the cell distribution between cell fractions helped confirm an initial difference between the unsorted and sorted fractions. At 7 days of culture, putative MPC colonies were enriched in the CD14-positive fraction ∼2-fold over unsorted cells and >20-fold over negatively selected cells. Over time, CD14 gene expression between unsorted and sorted cells was not different in cells cultured 14 days or more, suggesting that the MPCs growing from all 3 fractions were similar to each other. The difference between fractions appeared to be the yield and quantity of MPCs available for analysis at early time points (eg, at 14 days, few MPCs were present in the negative fraction in all samples). These results suggest that, in the horse, bone marrow-derived MPCs are not CD14 negative as reported in humans and other species.
In this study we have documented that CD14 protein and gene expression were both upregulated in equine MPCs after stimulation by LPS. Cells outside of the hematopoietic lineage normally do not express CD14 and should not respond to LPS stimulation. Therefore, either contaminating hematopoietic cells such as monocytes or dendritic cells are still present in MPC cultures at 21 days, or cultured MPCs do have low levels of CD14 expression on their cell surface, which allows them to respond to LPS treatment.
In the original characterization article by Pittenger et al., CD14 expression was reported as negative in human MPCs [6]. In that study, culture-expanded cells were harvested using either trypsin or EDTA, with no reported difference in cell surface protein detection for any marker on human MPCs between cell dissociation solutions. Most subsequent MPC characterization studies have used the protocol described by Pittenger, including trypsinization, for cell harvest before flow cytometric analysis. It is possible that trypsin has interfered with detection of the CD14 epitope on candidate cells analyzed in previous MPC studies.
Many reports in humans and other species consider CD14 negative, even though they document low levels of CD14 expression. For example, a recent MPC characterization study performed in sheep reported 28.5% (±13.8) of cultured MPCs were positive for CD14 expression at passages 3 and 4, but the authors still classified the ovine cells as CD14 negative [21]. The percentage of positive ovine MPCs in that study may have been artificially lowered since trypsin was used for cell harvest before flow cytometric analysis.
We have clearly demonstrated that equine MPCs had a low mean fluorescence intensity of CD14 detection when cells were treated with Accumax cell dissociation solution. When trypsin was used instead, CD14 expression was no longer detected in samples from the same MPC fractions. Our data suggest that CD14 is a trypsin-labile protein in the horse. Some additional equine cell surface proteins were also documented to be trypsin-labile, including CD44 and CD172a, whereas others such as CD90 had no detectable sensitivity to trypsin. Given these findings, the use of trypsin before flow cytometric analysis in equine bone marrow cells is not recommended.
Our results suggest that equine MPCs are enriched by selection for CD14 protein expression in bone marrow cells cultured for 2 days. Equine MPCs appear to have long-term expression of CD14 protein in cultured cells that can be upregulated by stimulation with LPS and damaged by exposure to trypsin, making them appear falsely negative for CD14. A plausible conclusion from these findings is that equine MPCs can be derived from a CD14-positive precursor cell. Ultimately, putative equine MPCs may represent a descendant population of a hematopoietic precursor.
Supplementary Material
Acknowledgments
The authors would like to express their gratitude to Bettina Wagner for providing the antiequine CD14 antibody used in this study. This study was funded by the Harry M. Zweig Memorial Fund for Equine Research Program, Grayson-Jockey Club Research Foundation and NIH T32RR07059 (C.H.H.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Kuwana M. Okazaki Y. Kodama H. Izumi K. Yasuoka H. Ogawa Y. Kawakami Y. Ikeda Y. Human circulating CD14+ monocytes as a source of progenitors that exhibit mesenchymal cell differentiation. J Leukoc Biol. 2003;74:833–845. doi: 10.1189/jlb.0403170. [DOI] [PubMed] [Google Scholar]
- 3.Kodama H. Inoue T. Watanabe R. Yasuoka H. Kawakami Y. Ogawa S. Ikeda Y. Mikoshiba K. Kuwana M. Cardiomyogenic potential of mesenchymal progenitors derived from human circulating CD14+ monocytes. Stem Cells Dev. 2005;14:676–686. doi: 10.1089/scd.2005.14.676. [DOI] [PubMed] [Google Scholar]
- 4.Pufe T. Petersen W. Fandrich F. Varoga D. Wruck CJ. Mentlein R. Helfenstein A. Hoseas D. Dressel S. Tillmann B. Ruhnke M. Programmable cells of monocytic origin (PCMO): a source of peripheral blood stem cells that generate collagen type II-producing chondrocytes. J Orthop Res. 2008;26:304–313. doi: 10.1002/jor.20516. [DOI] [PubMed] [Google Scholar]
- 5.Sera Y. Larue AC. Moussa O. Mehrotra M. Duncan JD. Williams CR. Nishimoto E. Schulte BA. Watson PM. Watson DK. Ogawa M. Hematopoietic Stem Cell Origin of Adipocytes. Exp Hematol. 2009;37:1108–1120. doi: 10.1016/j.exphem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pittenger MF. Mackay AM. Beck SC. Jaiswal RK. Douglas R. Mosca JD. Moorman MA. Simonetti DW. Craig S. Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Ringe J. Leinhase I. Stich S. Loch A. Neumann K. Haisch A. Haup T. Manz R. Kaps C. Sittinger M. Human mastoid periosteum-derived stem cells: promising candidates for skeletal tissue engineering. J Tissue Eng Regen Med. 2008;2:136–146. doi: 10.1002/term.75. [DOI] [PubMed] [Google Scholar]
- 8.Sung JH. Yang HM. Park JB. Choi GS. Joh JW. Kwon CH. Chun JM. Lee SK. Kim SJ. Isolation and characterization of mouse mesenchymal stem cells. Transplant Proc. 2008;40:2649–2654. doi: 10.1016/j.transproceed.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Han K. Lee JE. Kwon SJ. Park SY. Shim SH. Kim H. Moon JH. Suh CS. Lim HJ. Human amnion-derived mesenchymal stem cells are a potential source for uterine stem cell therapy. Cell Prolif. 2008;41:709–725. doi: 10.1111/j.1365-2184.2008.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You Q. Cai L. Zheng J. Tong X. Zhang D. Zhang Y. Isolation of human mesenchymal stem cells from third-trimester amniotic fluid. Int J Gynaecol Obstet. 2008;103:149–152. doi: 10.1016/j.ijgo.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Boiret N. Rapatel C. Veyrat-Masson R. Guillouard L. Guerin JJ. Pigeon P. Descamps S. Boisgard S. Berger MG. Characterization of nonexpanded mesenchymal progenitor cells from normal adult human bone marrow. Exp Hematol. 2005;33:219–225. doi: 10.1016/j.exphem.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Jo CH. Ahn HJ. Kim HJ. Seong SC. Lee MC. Surface characterization and chondrogenic differentiation of mesenchymal stromal cells derived from synovium. Cytotherapy. 2007;9:316–327. doi: 10.1080/14653240701291620. [DOI] [PubMed] [Google Scholar]
- 13.Maurice S. Srouji S. Livne E. Isolation of progenitor cells from cord blood using adhesion matrices. Cytotechnology. 2007;54:121–133. doi: 10.1007/s10616-007-9077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radcliffe CH. Flaminio MJ. Fortier LA. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2009;19:269–282. doi: 10.1089/scd.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryniarski K. Maresz K. Szczepanik M. Ptak M. Ptak W. Modulation of macrophage activity by proteolytic enzymes. Differential regulation of IL-6 and reactive oxygen intermediates (ROIs) synthesis as a possible homeostatic mechanism in the control of inflammation. Inflammation. 2003;27:333–340. doi: 10.1023/b:ifla.0000006701.52150.43. [DOI] [PubMed] [Google Scholar]
- 16.Flaminio MJ. Rush BR. Shuman W. Peripheral blood lymphocyte subpopulations and immunoglobulin concentrations in healthy foals and foals with Rhodococcus equi pneumonia. J Vet Intern Med. 1999;13:206–212. doi: 10.1892/0891-6640(1999)013<0206:pblsai>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Kabithe E. Hillegas J. Stokol T. Moore J. Wagner B. Monoclonal antibodies to equine CD14. Vet Immunol Immunopathol. 2010. [Epub ahead of print]. [DOI] [PubMed]
- 18.Mauel S. Steinbach F. Ludwig H. Monocyte-derived dendritic cells from horses differ from dendritic cells of humans and mice. Immunology. 2006;117:463–473. doi: 10.1111/j.1365-2567.2005.02319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 20.Goidin D. Mamessier A. Staquet MJ. Schmitt D. Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295:17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 21.McCarty RC. Gronthos S. Zannettino AC. Foster BK. Xian CJ. Characterisation and developmental potential of ovine bone marrow derived mesenchymal stem cells. J Cell Physiol. 2009;219:324–333. doi: 10.1002/jcp.21670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.