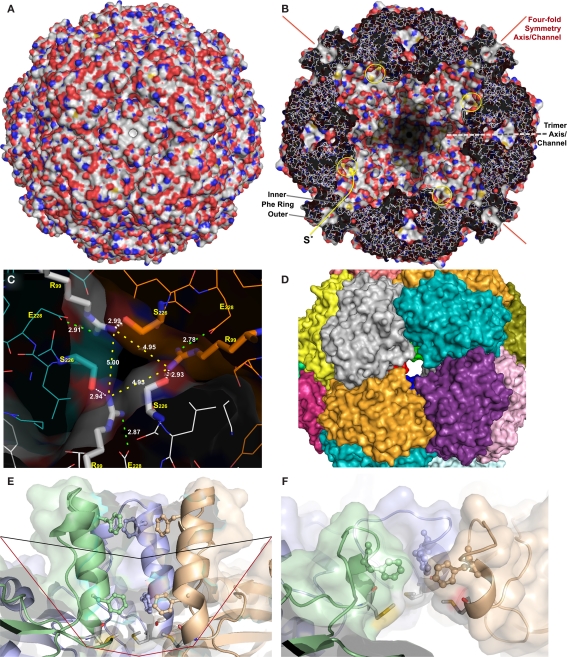
Figure 2.
X-ray crystallography and modeling of the SOR and its pores. (A) Surface representation of the holoenzyme centered at the chimney-like structure at the fourfold symmetry axis. (B) Representation of the protein structure and the inner surface of the holoenzyme sliced at the center of the fourfold symmetry axes; the position of the inner and outer phenylalanine rings are indicated, also the active site pores (yellow circles), and the approximate position of the trimer symmetry axis, which is tilted out of plane. (C) Channel at the threefold symmetry axis formed by R99 and S226; distances are given between the Nη atoms of the arginines (yellow dashes), for the salt bridges to E228 (green dashes), and for the putative hydrogen bond to the Oγ of the S226 of the neighboring subunit (white dashes). (D) Subunit representation of the large deletion mutant at the fourfold symmetry axis DelL (Figure 1) modeled at the SwissModel server (Arnold et al., 2006). (E) Side view of the channel at the fourfold symmetry axis with outer (F141) and inner phenylalanine rings (F133; from top, showing three out of four subunits; wild type) and the methionine ring at its base (M130); black line, approximate position of small deletion (DelK); red line, large deletion of entire chimney (DelL). (F) Model of the same three subunits as in panel E of the short DelK deletion, modeled at the Phyre server (Kelley and Sternberg, 2009).