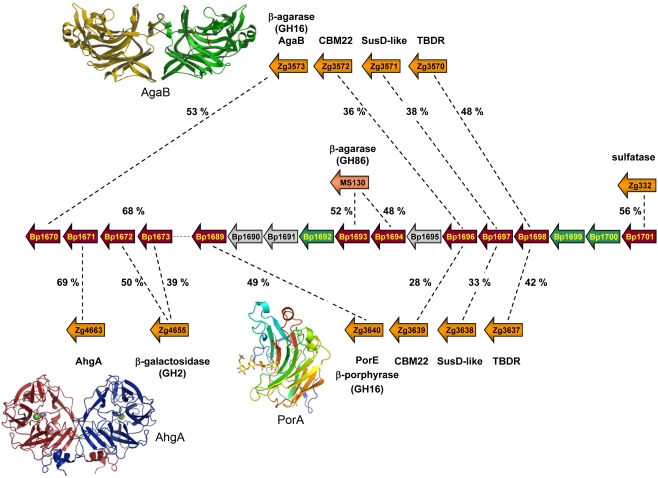
Figure 2.
Schematic representation of a PUL region, present in the genome of Bacteroides plebeius and first identified as interesting because of the presence of a porphyranase gene (Bp1689; Hehemann et al., 2010). Besides the conserved Sus-like genes, the locus also contains carbohydrate-related genes which share highest identity with proteins used for red algal galactan degradation in two marine Bacteroides. Shown are the sequence identities between B. plebeius and one Microscilla sp. PRE1 protein, as well as with several Zobellia galactanivorans proteins. Six of these genes (Bp1670, Bp1671, Bp1689, Bp1693, Bp1694, and Bp1696) are conserved only with marine bacteria, and are absent in genomes of other gut Bacteroides. The crystal structures of marine homologous enzymes, coded by three of these genes (namely Bp1670, Bp1671, and Bp1689) have recently been determined and are illustrated as ribbon representations (PDB codes: AgaB – 1O4Z; AghA – 3P2N; and PorA – 3ILF). The 3D structures help determine the crucial residues for activity and substrate specificity that are all verified and present in the sequences of the B. plebeius proteins.