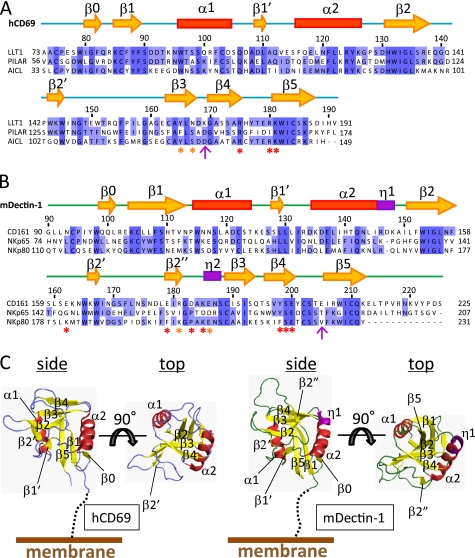
FIGURE 2.
Amino acid sequence alignment and structure of CTLDs. Shown are amino acid sequence alignments of the CTLDs of LLT1 with PILAR and AICL (A) and CD161 with NKp65 and NKp80 (B). Secondary structure elements (yellow arrows indicate β-strands, red boxes indicate α-helices, and purple boxes indicate 310 helices) of hCD69 and mDectin-1 are displayed above the alignments. The asterisks indicate residues mutated in this study, with detrimental effect in red and modest effect in orange. The magenta arrows indicate the pair of residues that showed detrimental effects when mutated independently but restored the binding when mutated simultaneously. C, ribbon diagrams of hCD69 (left) and mDectin-1 (right). Each diagram shows side and top views. Secondary structure elements are labeled as follows: α-helices (red), β-strands (yellow), and 310 helices (purple).