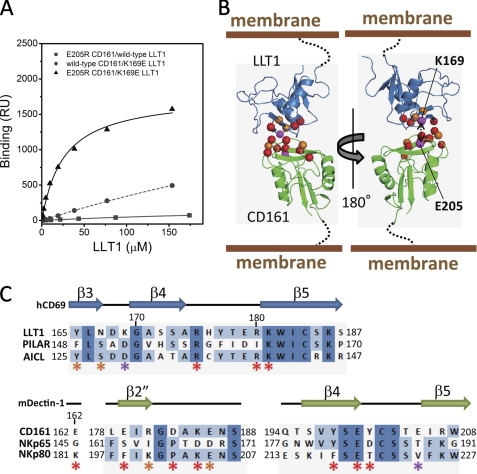
FIGURE 4.
Model structure of CD161-LLT1 complex. A, SPR binding analyses of combinations between the LLT1 and CD161 mutants. Filled squares and solid line, E205R CD161/wild type LLT1; filled circles and dashed line, wild type CD161/K169E LLT1; filled triangles and solid line, E205R CD161/K169E LLT1. B, schematic model of LLT1 recognition by CD161. Ribbon diagrams and spheres are the same as in Fig. 3C. C, amino acid sequence alignments of the putative receptor/ligand binding regions of LLT1 with PILAR and AICL (top) and CD161 with NKp65 and NKp80 (bottom). Secondary structure elements of hCD69 and mDectin-1 are displayed above the alignments. The asterisks indicate residues mutated in this study, with color representations corresponding to those in B and Fig. 3C.