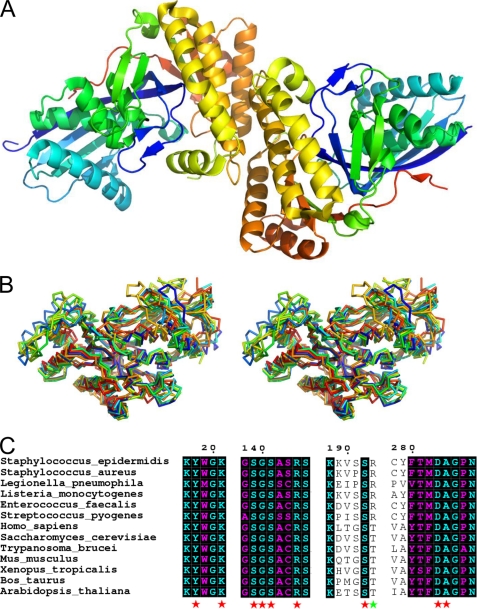
FIGURE 2.
1.85 Å crystal structure of apo-MDD from S. epidermidis. A, crystal structure of S. epidermidis MDD shown in ribbon format. Two copies of MDD are found within the asymmetric unit and are colored blue (N terminus) and red (C terminus). B, stereo view of MDD structures in ribbon format deposited within the PDB. Structures correspond to the following organisms: S. epidermidis (red-orange); S. aureus (cyan); L. pneumophila (orange); S. pyogenes (red); H. sapiens (yellow-green); S. cerevisiae (blue); T. brucei (green); and M. musculus (yellow). C, limited structure-based sequence alignment of prokaryotic and eukaryotic MDD proteins. Alignment was generated using ClustalW and rendered with ESPRIPT. Numbers above the sequences correspond to S. epidermidis MDD. Red stars below the sequences correspond to invariant amino acid side chains involved in DPGP and FMVAPP interaction, and the green star represents the single variable active site residue. Sequences from B as well as the following were used in alignment: L. monocytogenes; E. faecalis; Xenopus. tropicalis; B. taurus; A. thaliana.