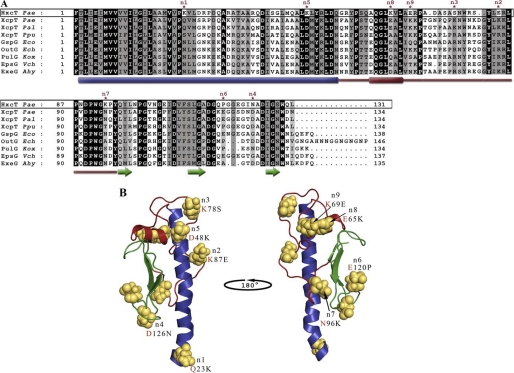
FIGURE 2.
Selection criteria for mutations of HxcT. A, sequence alignment of matured major pseudopilins from T2SSs. Sequence of HxcT from the T2SS of P. aeruginosa PAO1 is boxed. Identical residues are highlighted in black, conserved residues are highlighted in dark gray, and similar residues are highlighted in light gray. The secondary structure elements are indicated below the sequence alignment where cylinders represent the α-helices, arrows represent the β-strand, and the red line indicates the αβ-loop region. The point mutations selected to transform HxcT in XcpT are indicate by asterisks. Pseudopilin gene identification (gi) numbers: Pal, P. alcaligenes M-1(gi 3978481/AAC83358.1); Ppu, P. putida WCS358 (gi 3297910/CAA56982.1); Eco, E. coli SMS-3-5 (gi 170681001/YP_001745224.1); Ech, E. chrysanthemi (gi 259985/AAB24181.1); Kox, K. oxytoca UNF5023 (gi 131595/P15746.1); Vch, V. cholerae MZO-3 (gi 153802200/ZP_01956786); Ahy, A. hydrophila ATCC 7966 (gi 117620343/YP_855105); and Pae, P. aeruginosa PAO1 (gi 15598297/NP_251791 and 15595878/NP_249372) for XcpT and HxcT, respectively. B, ribbon view of the XcpT pseudopilin from P. aeruginosa (PAO1). N-terminal helix is colored in blue, αβ-loop region in red, and the C-terminal three-stranded β-sheet is colored in green. Residues selected to transform HxcT into XcpT are represented in sphere mode. Mutations performed are indicated below the priority tag with a red letter for the XcpT residue.