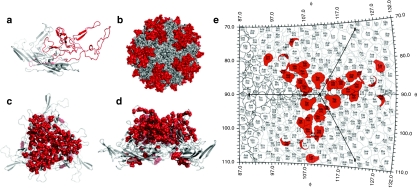
Figure 1.
Structural analysis of the adeno-associated virus serotype 9 (AAV9) capsid library. (a) Cartoon representation of the AAV9 VP3 subunit monomer obtained using SWISS-MODEL with crystal structure of AAV8 serving as template (pdb id: 2QA0). The GH loop containing amino acids 390–627 (VP1 numbering) is colored in red. (b) Surface rendering of an AAV9 capsid model with 60 VP3 subunits generated using T = 1 icosahedral symmetry coordinates on VIPERdb. GH loop regions from different VP3 subunits, surrounding the icosahedral fivefold pore and interdigitating at the threefold symmetry axis are highlighted in red. (c) Cartoon of AAV9 VP3 subunit trimer generated on VIPERdb with point mutations of 43 representative clones from the AAV9 library depicted by red spheres. (d) Side view of capsid trimer (90° rotation) showing a majority of point mutations (red spheres) clustered on the outer loops. (e) Spherical roadmap projection of surface residues within the capsid trimer region. Residues highlighted in red represent a subset of ten AAV9 variants containing altered residues prominently located on the capsid surface.