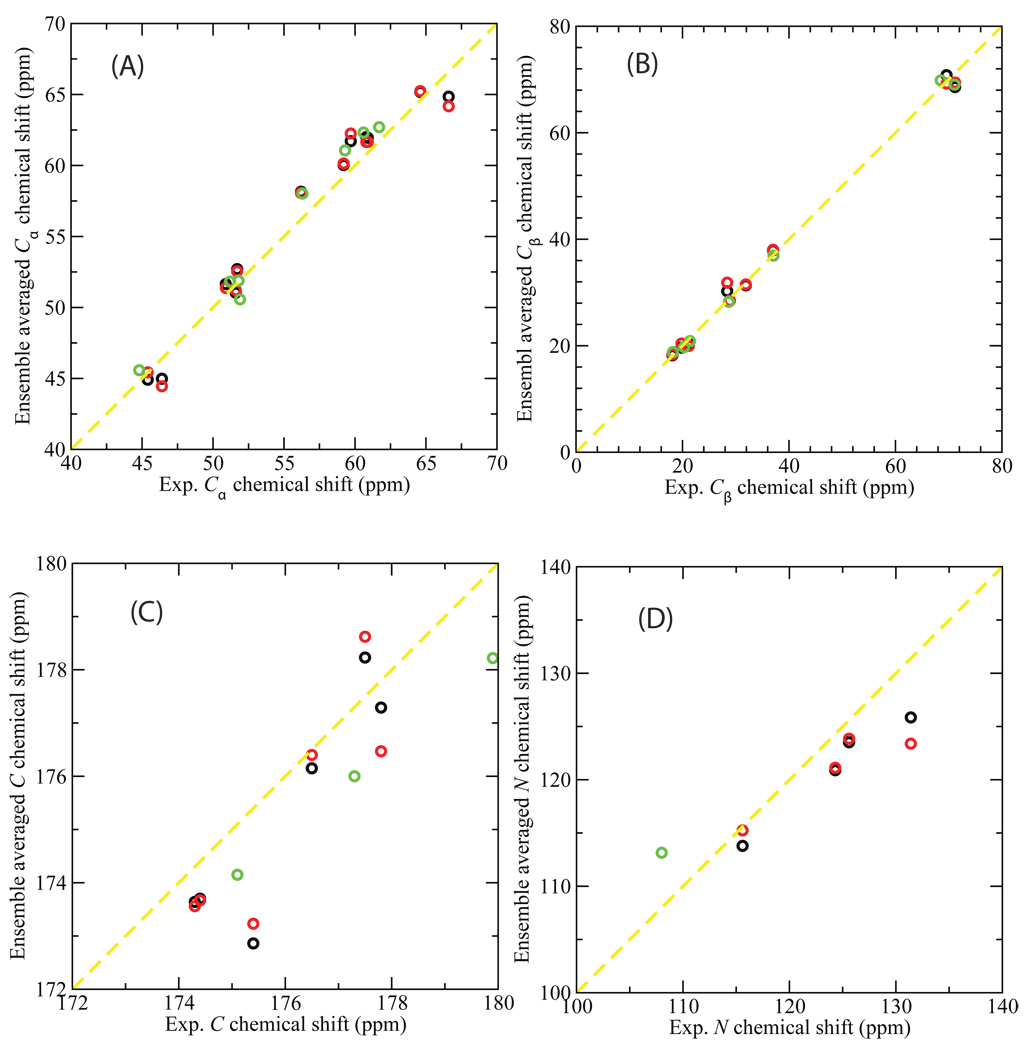
Fig. 6.
Ensemble-averaged chemical shifts (ppm) versus the NMR experimental measurements for Cα (A), Cβ (B), carbonyl C (C), and amide N (D) atoms of the flexible loop 6 of yeast TIM. SHIFTX56 was used to calculate chemical shifts which were then averaged over an ensemble of 1000 structures from the equilibrated MC simulations. The starting PDB structures for the simulations are: 1YPI (black); 2YPI with the ligand PGA removed (red); and 2YPI with PGA bound (green). The experimental chemical shift data are those for apo yeast TIM in NMR experiment57 (for comparison with the apo simulations), and for yeast TIM with ligand G3P57 (for comparison with the holo simulation). Experimental chemical shifts are not available for some atoms and these are omitted.