Abstract
PHD (plant homeodomain) zinc fingers are structurally conserved modules found in proteins that modify chromatin as well as mediate molecular interactions in gene transcription. The original discovery of their role in gene transcription is attributed to the recognition of lysine-methylated histone H3. Recent studies show that PHD fingers have a sophisticated histone sequence reading capacity that is modulated by the interplay between different histone modifications. These studies underscore the functional versatility of PHD fingers as epigenome readers that control gene expression through molecular recruitment of multi-protein complexes of chromatin regulators and transcription factors. Moreover, they reinforce the concept that evolutionary changes in amino acids surrounding ligand binding sites on a conserved structural fold impart great functional diversity upon this family of proteins.
The PHD finger as a sequence-specific histone recognition protein module
Both gene transcriptional activation and silencing are directed by post-translational amino acid modifications to the DNA-packing core histones, H2A, H2B, H3, and H4 [1–3]. Of the known histone modifications, acetylation of lysine, and methylation of lysine or arginine play a direct role in orchestrating the recruitment of multi-protein complexes for changing chromatin structure at target gene loci, for processing gene transcriptional initiation, elongation and termination, as well as for transcriptional repression [4–7]. These histone sequence- and state-specific events are manifested through molecular interactions with structurally conserved protein modular domains that are present in many chromatin regulators and transcription factors. The cross-talk and molecular interplay between these different modifications that take place in relatively short stretches of nucleosomal histone tails highlight the delicate and complex nature of gene regulation in chromatin between gene activation and silencing states.
Several protein modular domains have been reported to recognize histones in a sequence- and modification-dependent manner. For example, bromodomains are known to function as acetyl-lysine binding domains in all eukaryotes [6, 8], whereas chromodomains and other members of their structurally related Royal family protein domains that also include the Tudor, MBT (malignant brain tumor), PWWP (Pro-Trp-Trp-Pro) and Agenet domains bind histones H3 and H4 in a methylation-sensitive manner [7, 9–14]. More strikingly, recent studies have illuminated that PHD (plant homeodomain) fingers [15, 16], composed of a small zinc finger structural fold, appear to have evolved the ability to read histone states in a sequence-dependent manner that is modulated positively or negatively by the methylation state of lysine and arginine [17–22], as well as by the acetylation state of lysine [23, 24]. In this article, we will review recent advancements in our knowledge of the structure and function of PHD fingers and discuss the functional versatility of this small conserved protein domain in the regulation of gene transcription.
Ligand recognition
The PHD finger is a small protein domain of 50–80 amino acid residues of diverse sequences containing a zinc-binding motif that appears in many chromatin-associated proteins [15] (Figure 1A). The conserved PHD fold consists of a two-strand anti-parallel β-sheet and a C-terminal α-helix (not present in all PHDs) that is stabilized by two zinc atoms anchored by the Cys4-His-Cys3 motif in a cross-brace topology [17, 25] (Figure 1B). PHD fingers can read the N-terminal tail of histone H3, mainly the methylation state of H3K4 (K4me0 vs K4me3/2) [17, 20, 26–31], and to a smaller extent the methylation state of H3R2 (R2me0 vs R2me2) [32–36] and the acetylation state of H3K14 [24]. Binding of yeast PHD fingers to methylated H3K36 has also been reported [37]. The reading of H3K4 and H3R2 occurs on a single PHD finger, but at distinct sites. By contrast, the single example of H3K14 reading occurs in conjunction with the reading of H3K4 and H3R2, in a tandem PHD finger of DPF3b (D4, zinc and double PHD fingers, family 3 protein), where the first PHD reads H3K14ac, and the second PHD reads H3K4me0 and H3R2me0 [24].
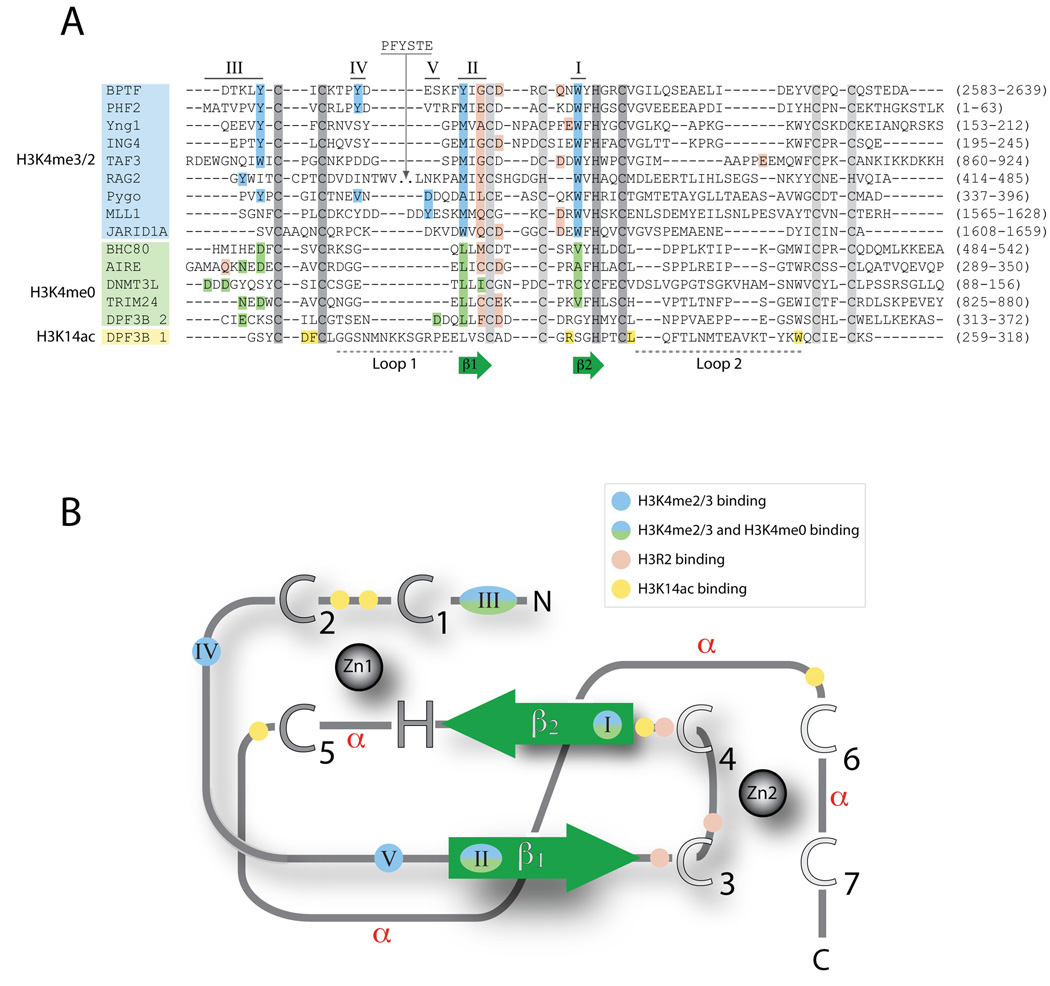
Figure 1. Conserved features of PHD fingers.
PHD fingers exhibit high sequence variability, but share some common sequence and structure patterns. Common features include the Zinc-coordinating residues, two core β-strands (β1 and β2), and the use of common sequence regions to form ligand binding sites. (A) Structure-based alignments of representative PHD finger sequences. The conserved Zinc-coordinating residues are shown in dark gray for Zinc 1 and light gray for Zinc 2, and the two core β-strands are shown in green. The regions involved in ligand recognition are labeled I through V, ordered by the frequency with which they are utilized within the domain family. Residues are colored according to their participation in the recognition of H3 tail residues: blue, H3K4me3 recognition; green, H3K4me0 recognition; pink, H3R2 recognition; and yellow, H3K14ac recognition. The residue numbers corresponding to the PHD finger in the full-length protein are shown in parenthesis. (B) Schematic of the PHD fold. All PHD fingers adopt the same basic topology. Zinc atoms are shown as gray spheres, with Zinc-coordinating residues indicated as large uppercase letters. The two core β-strands are shown in green. The regions involved in ligand recognition are labeled (I–V) and colored as indicated in A. Regions that adopt an α-helical conformation in some PHD fingers are indicated (α).
The first structure of a PHD finger bound to its ligand was that of bromodomain PHD finger transcription factor (BPTF) [17] which binds the H3 tail when K4 is trimethylated (H3K4me3) and R2 is unmodified (H3R2me0). The BPTF structure revealed several features that are common to most PHD fingers. The most conserved feature is the binding of the first six N-terminal residues of H3 (ARTKQT) on the surface of the PHD finger through the formation of an anti-parallel β-strand to the two-strand β-sheet (β1 and β2) of the PHD finger (Figure 2). The backbone amide and carbonyl groups of residues R2, T3, and K4 of H3 form hydrogen bonds with the β1 strand backbone. This β-strand conformation orients R2 and K4 into two distinct pockets separated by a highly conserved Trp residue (position I in Figure 1A), while the N-terminal A1 residue is stabilized by interactions with residues in Loop 2 and/or residues found between the β1 and β2 strands. In general, binding of the H3 tail does not seem to induce significant conformational changes in the PHD finger [17, 20].
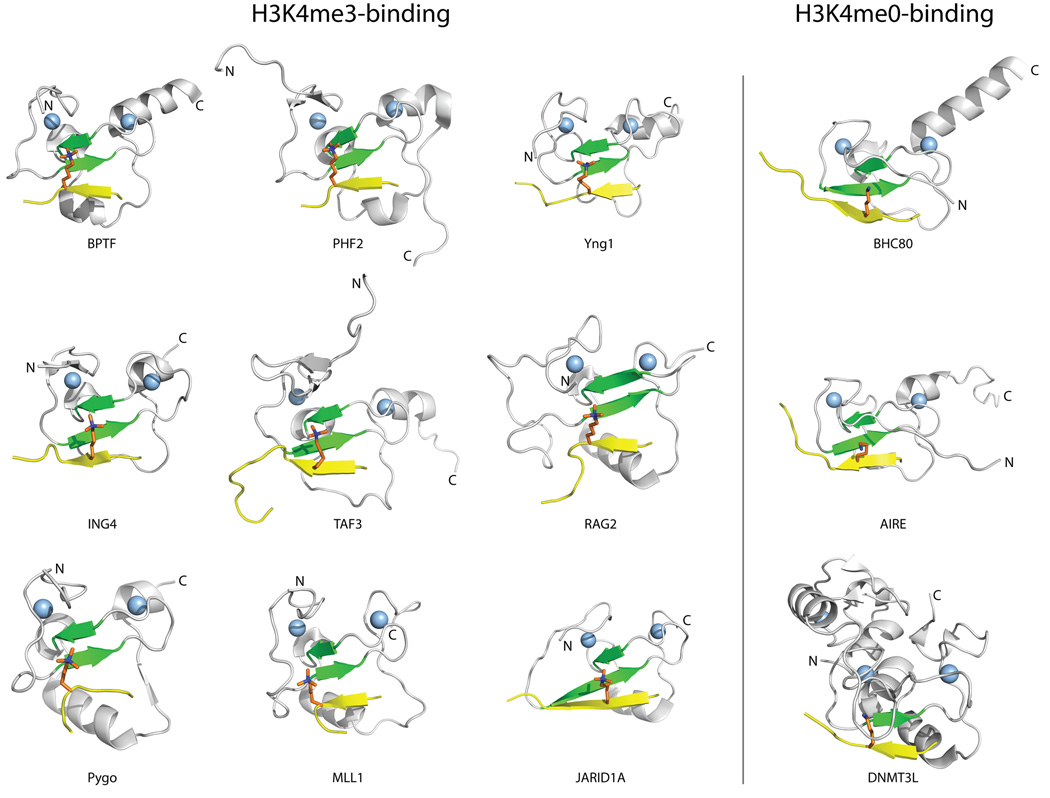
Figure 2. Structures of representative PHD fingers in complex with methylated or unmethylated H3K4 peptides.
The common elements in the binding of PHD fingers to H3 histone tails are illustrated for PHD fingers that bind H3K4me3 (i) and H3K4me0 (ii). The PHD finger structures are shown in cartoon representation with the conserved β-strands (β1 and β2) colored green, and the Zinc atoms represented by light blue spheres. The H3 peptide ligand (shown in yellow, with the K4 residue colored orange) adopts a β-strand conformation that extends the conserved anti-parallel β-sheet formed by β1 and β2 in the PHD finger. The following PDB structures were used to create this figure: 2f6j (BPTF), 3kqi (PHF2), 2jmj (Yng1), 2pnx (ING4), 2k17 (TAF3), 2v83 (RAG2), 2yyr (PYGO), 3lqj (MLL1), 2kgi (JARID1A), 2puy (BHC80), 2ke1 (AIRE), and 2pvc (DNMT3L).
Reading H3K4me3
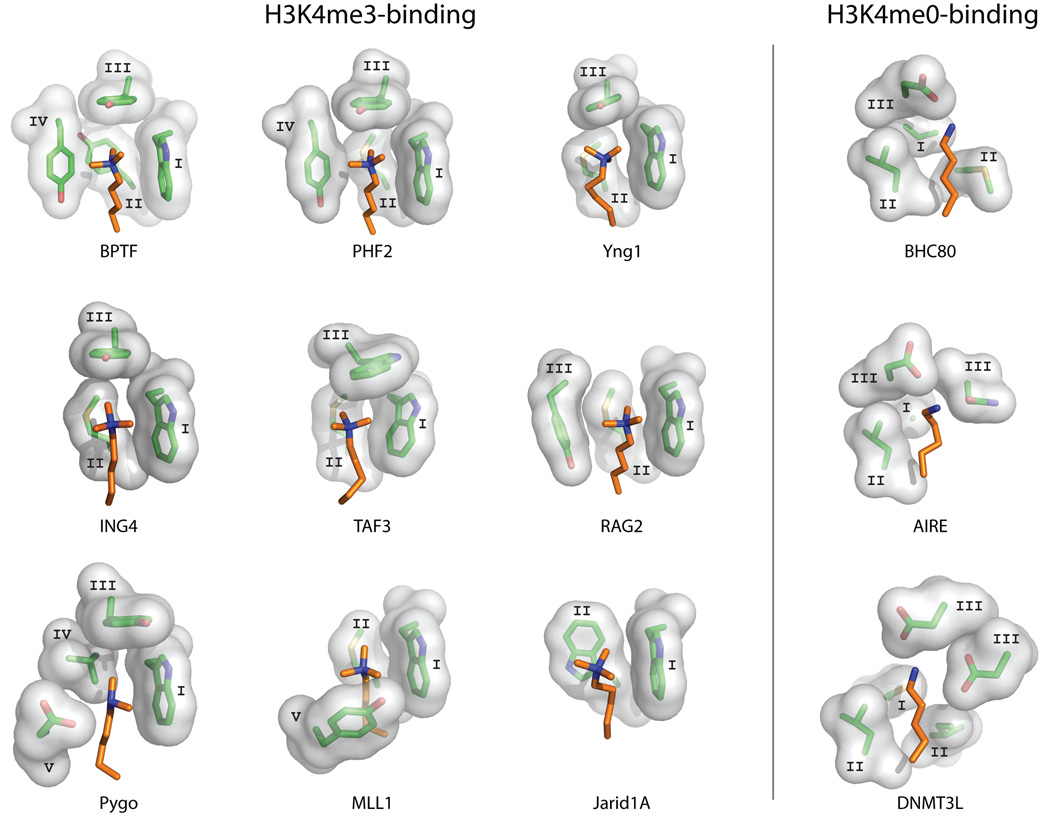
The BPTF-PHD structure also revealed the main characteristics of PHD fingers that read H3K4me3. The binding of H3K4me3 occurs through an aromatic cage (Figure 3) where the trimethyl ammonium group is stabilized by van der Waals and cation-π interactions, similar to those observed in chromodomain, MBT, PWWP and Tudor domains [11, 14, 38–40]. In BPTF this aromatic cage is composed of one Trp and three Tyr residues; the cage has three faces and a “lid” beyond the tip of H3K4me3 (Figure 3). Subsequently determined structures of other PHD fingers in complex with H3K4me3 peptides show that the aromatic cage varies, containing a combination of 2 to 4 aromatic and hydrophobic residues (Figure 3). The residues participating in the aromatic cage tend to appear at similar positions in the sequence (Figure 1). The most conserved position is an invariable Trp residue at the beginning of the β1 strand (Position I in Figure 1), followed by an aromatic or hydrophobic residue at the start of β2 (Position II in Figure 1). In general, residues that are used to form the aromatic cage tend to exist in parts of the structure that are relatively rigid, such as the β strands, or close to the Zn-coordinating Cys residues (positions I–V in Figure 1). A minimum of two aromatic residues including the invariable Trp at position I (Figure 1A) seem to be required for H3K4me3 binding. The simplest aromatic cage observed so far is that of jumonji, AT-rich interactive domain 1A (JARID1A) [26], which is composed of only two Trp residues at positions I and II (Figure 3). Together with JARID1A, the aromatic cages of recombination activating gene 2 (RAG2) and myeloid/lymphoid or mixed-lineage leukemia-1 (MLL1) lack the “lid” residue at position III present in all other PHD finger aromatic cages (Figure 3). Additionally, Y1581 (position V) in MLL1 undergoes a conformational change upon binding (together with the rest of the Y1576-Met1585 loop), which is not observed in other PHD fingers. A slight variation in the H3K4 binding region is observed in the PHD fingers of yeast homolog of mammalian ING1 (Yng1), transcription initiation factor TFIID subunit 3 (TAF3), pygopus homolog 1 (PYGO), inhibitor of growth protein 4 (ING4), and other PHD fingers of the ING family that have charged (Asp) or hydrophilic (Ser) residues in close proximity to the H3K4 residue, at position IV or V (Figure 1A). Of these, only the Asp residue at position V in PYGO plays a role in methylated lysine binding [20, 27–29], by slightly shifting the affinity in favor of the dimethylated form of K4 (H3K4me2), making the affinities of the free PHD for H3K4me3 (KD=2.5µM) and H3K4me2 (KD=2.4µM) virtually identical, and showing a slight preference for H3K4me2 (KD=0.9µM) over H3K4me3 (KD=1.2µM) and H3K4me1 (KD=2.9µM) in the PHD–HD1 complex [29]. Although the Ser and Asp residues in the other PHDs contribute to H3 binding, they do so via interactions with residues other than K4 [29]. Mutational studies in BPTF also suggest that the presence of a negatively charged residue in the aromatic box can alter the binding selectivity of the PHD finger [41]. The affinities in the wild-type BPTF, which show a preference for H3K4me3 (KD=3µM) over H3K4me2 (KD=6µM), change upon mutation (generating in a substitution of Tyr to Glu at position IV) resulting in a preference of H3K4me2 (KD=7µM) over H3K4me3 (KD=15µM) [41]. Thus, it seems that the relative affinity of H3K4me3 vs. H3K4me2 can be modulated by subtle changes in the sequence of the PHD finger. However, the biological impact of these small differences in affinity is not clear.
Figure 3. Ligand recognition sites in PHD fingers recognizing H3K4me3 or H3K4me0.
The binding sites are illustrated by showing the sidechains of the PHD finger residues (stick and surface representation) that surround the H3K4 ligand residue (shown in orange). (i) The different versions of aromatic cages used for recognition of H3K4me3 are shown. (ii) H3K4me0 binding sites are shown. The binding site residues are labeled according to the sequence regions defined in Figure 1. The PDB structures used to create this figure are listed in Figure 2.
Recently, the structure of a zinc finger CW (zf-CW) domain in complex with an H3K4me3 peptide was reported [42]. The zf-CW domain adopts a fold in which a zinc ion is coordinated by four conserved Cys ligand residues, which resembles one half of a PHD finger. Additionally, an aromatic cage formed by the same conserved positions used by H3K4me3-recognizing PHD fingers (Figure 1) mediates the recognition of H3K4me3 in zf-CW. Hence, it is likely that zf-CW domains and PHD fingers share a common evolutionary ancestor.
Reading H3K4me0
A separate group of PHD fingers, which includes those of BRAF35-HDAC complex protein (BHC80), autoimmune regulator (AIRE), tripartite motif-containing protein 24 (TRIM24), and DNA (cytosine-5)-methyltransferase 3-like protein (DNMT3L), lack an aromatic cage [32–36, 43]. These PHD fingers bind unmodified H3K4 (H3K4me0), and this interaction is abrogated by the methylation of H3K4 [32–36, 43]. Similar to the H3K4me3-binding PHD fingers, the histone H3 tail also forms a β-sheet with the β strands in the H3K4me0-binding PHD fingers (Figure 2). The binding of K4me0 is achieved by a combination of acidic and hydrophobic residues that replace the aromatic cage (Figure 3). Indeed, the same sequence regions used to form the aromatic cage in H3K4me3-binding PHD fingers, corresponding to structurally stable positions, are used to form the binding site in H3K4me0-binding PHD fingers (regions I–III in Figure 1A & B).
Reading H3R2
When bound to the PHD fingers of BPTF, Yng1, ING4, TAF3, MLL1, or JARID1A, the side chains of H3R2 and H3K4me3 extend into two adjacent binding pockets separated by the conserved Trp residue at position I (Figure 4A) [17, 20, 26–28, 44]. In these cases, the guanidinium group of H3R2 forms salt bridges or hydrogen bonds with Asp, Glu and Gln residues. Additionally, the volume of the pocket is maintained by the presence of a small residue (Gly or Ala) at the position preceding the third Zn-coordinating Cys residue (in BPTF, Yng1, ING4, and TAF3) [17, 20, 27, 28]. Alternatively, the pocket can be shifted by the presence of a Gln residue at the same position, which stacks against the conserved Trp residue at position I (in MLL1 and JARID1A) [26, 44]. In all of these cases, the binding pocket makes favorable interactions with unmodified H3R2 (H3R2me0), and affinity for the H3 tail decreases when R2 is mono- or dimethylated [17, 20, 26–28, 44]. A variation on this theme is observed in PHD finger protein 2 (PHF2) in which a proper pocket for H3R2 binding is not observed, but a Glu at the position preceding the third Zn-coordinating Cys residue enables favorable interactions with H3R2me0 [31]. Binding of the H3 tail to PHD1 of AIRE is also antagonized by H3R2 methylation [36]. Again, the guanidinium group of H3R2 forms a salt bridge with Asp 312, which is disrupted upon methylation of H3R2. Hence, PHD1 of AIRE reads a completely unmodified H3 tail (R2me0 + K4me0).
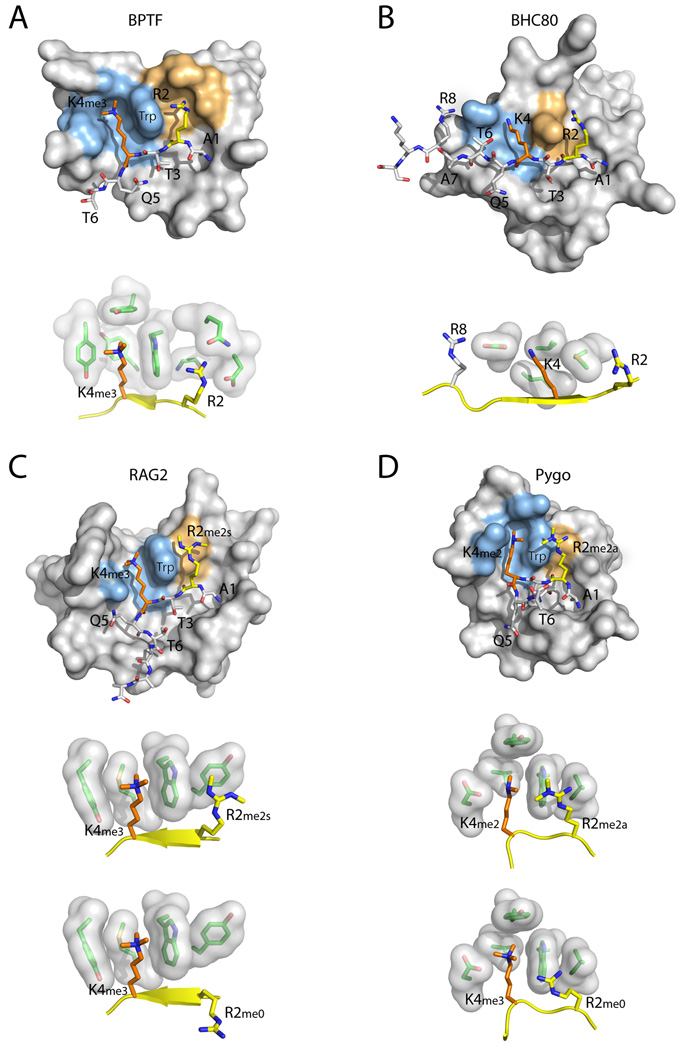
Figure 4. Recognition of multiple H3 residues by PHD fingers.
The structures of representative PHD fingers bound to H3 peptides with different combinations of H3K4 and H3R2 methylation states are shown. The close-up images show the residues involved in recognition of H3K4 and H3R2. (A) BPTF PHD bound an H3 R2me0;K4me3 peptide. (B) BHC80 PHD bound to an H3 R2me0;K4me0 peptide. (C) RAG2 PHD bound to an H3 R2me2s;K4me3 peptide (the bottom image shows an H3 R2me0;K4me3 peptide). (D) PYGO PHD bound to an H3 R2me2a;K4me2 peptide (the bottom image shows an H3 R2me0;K4me3 peptide). The H3 N-terminal peptide is shown in stick representation, with H3K4 in orange and H3R2 in yellow. The H3K4 recognition environment is colored light blue on the surface of the PHD finger, and the H3R2 recognition environment is colored orange. In the close-up images, the H3 peptide is shown in yellow cartoon representation.
A different situation is observed in RAG2 and PYGO. RAG2 PHD has a slight preference for symmetrically dimethylated R2 (R2me2s, KD=25.1µM) over unmethylated (R2me0, KD=33.8µM), or asymmetrically dimethylated R2 (R2me2a, KD=34.6µM) [30]. In this domain, a Tyr residue is observed at the position that is usually occupied by a small residue in the other PHDs (Figure 1A). This Tyr residue provides an interacting surface for R2me2s (Figure 4C). Together with the absence of nearby acidic residues, this change seems to favor binding of R2me2s over R2me0, R2me1, or R2me2a [30]. Although the biological significance of this dual mark is not clear, RAG2-PHD might be an example of a single protein domain recognizing a doubly modified H3 histone tail (R2me2s + K4me3) (Figure 4C). At the very least, RAG2-PHD differs from the other PHDs in that its binding to H3K4me3 is not diminished by methylation of H3R2. A similar situation is observed in PYGO; the same position occupied by the Tyr residue in RAG2 is occupied by Leu (Figures 1A and 4D). This change abolishes the H3R2me0 binding pocket; however, it does not favor H3R2me2 or H3R2me1 binding, thus rendering PYGO insensitive to H3R2 methylation [29]. Hence, it appears that depending on the residues present at the position preceding the third Zn-coordinating Cys residue, and the presence or absence of nearby acidic residues, a PHD finger can prefer to bind H3R2me0, H3R2me2, or it can be insensitive to the H3R2 methylation state.
Reading H3K14ac
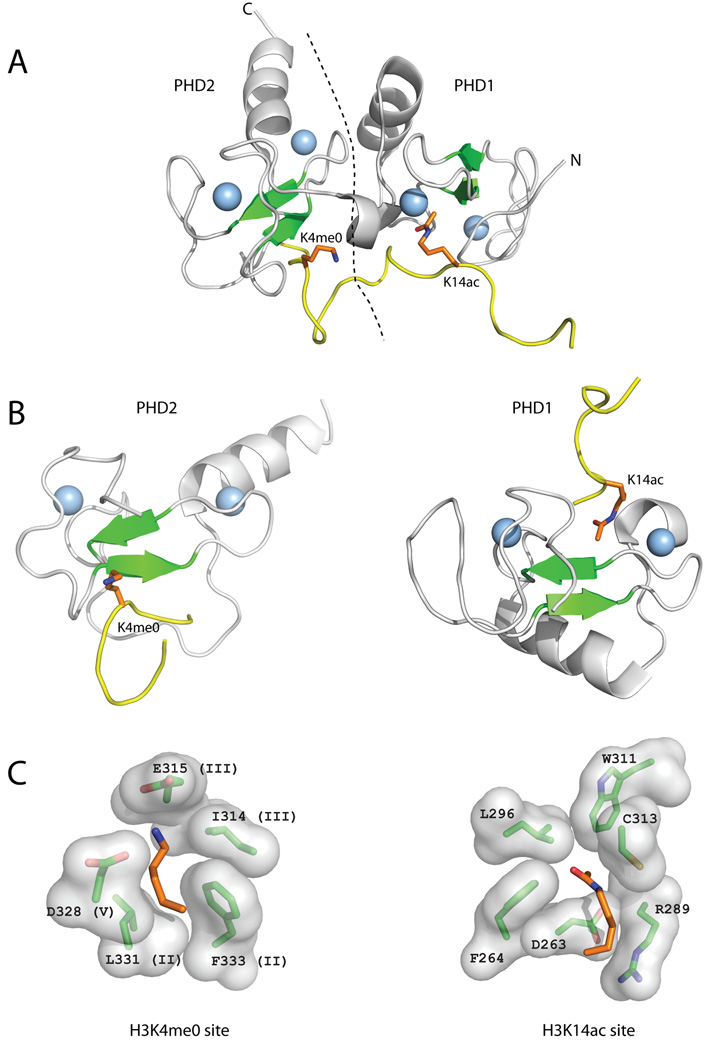
Recently, the tandem PHD finger of DPF3b was shown to bind H3 acetylated at K14 (H3K14ac)[24]. This is the first, and so far only, example of a PHD finger recognizing an acetylated lysine, a post-translational modification usually recognized by bromodomains [6, 8]. The first PHD finger (PHD1) in the DPF3b tandem binds H3K14ac, whereas the second PHD finger (PHD2) binds the H3R2-K4 regions (Figure 5). DPF3b-PHD2 is similar to the H3K4me0-binding PHD fingers (BHC80, AIRE, DNMT3L): it shares their combination of acidic and hydrophobic residues in the H3K4me0 binding pocket (Figures 1A & 5). The H3K14ac binding pocket in DPF3b-PHD1 is, however, completely novel. It is located on the opposite side of the central β-sheet and uses a set of residues that do not overlap with the positions used to form the H3K4 or H3R2 binding sites (Figures 1 & 5). Nevertheless, the H3K14ac binding site follows the same principle of using structurally rigid positions that are near either Zn-coordinating Cys residues or the β2 strand (Figure 1B).
Figure 5. Reading of H3 tail by the DPF3b tandem PHD fingers.
The DPF3b tandem PHD fingers bind an H3 peptide acetylated on K14 (H3K14ac) and unmodified at R2 (H3R2me0) and K4 (H3K4me0) (A) The structure of the DPF3b tandem PHD fingers in complex with the histone H3 N-terminal tail peptide (PDB code 2kwj) is shown in cartoon representation (coloring as in Figure 2). The dotted line represents the boundary between PHD1 and PHD2. (B) The structures of PHD1 and PHD2 of DPF3b bound to H3K4me0 and H3K14ac, respectively, are shown separately in the same orientation and coloring scheme as the PHD fingers in Figure 2. (C) Close-up images of the H3K4me0 and H3K14ac recognition sites showing the residues involved in binding labeled according to residue number, and sequence region (for H3K4me3) (Figure 1).
H3 specificity
The PHD fingers that have been structurally characterized illustrate several elements that contribute to specificity in binding to the histone H3 N-terminal tail. As mentioned above, the N-terminal A1 residue is stabilized by interactions with residues in Loop 2 and/or residues found between the β1 and β2 strands. In addition to the specific environments that read the methylation state of H3R2 and H3K4, the spacing between these residues, provided by H3T3, strongly contributes to the binding specificity. A highly conserved Trp residue (position I) occupies the space between H3R2 and H3K4 in the H3K4me3-binding PHD-fingers (Figure 4), and the deletion of T3 abrogates binding [4]. In H3K4me0-binding PHD fingers, the space between H3R2 and H3K4 is occupied by hydrophobic residues such as Met, Ile, and Phe at position II in BHC80, DNMT3L, and DPF3b-PHD2, respectively, and Asn at position III in AIRE (Figure 1A). This spacing requirement prevents the PHD fingers from reading, for example, methylated H3K9 or H3K27, because both sites are immediately preceded by an arginine. It is worth noting, however, that methylated H3K36, which is preceded by a Val residue, has been reported to be recognized by yeast PHD fingers [37]. Although it is possible that among the large number of PHD fingers some exhibit a different histone-binding specificity, this has not been observed so far, and would require changes to the PHD finger sequence.
Combinatorial Reading
The ability of PHD fingers to read the modification state of H3R2, H3K4, and H3K14 enables the possibility of identifying combinations of modifications by using more than one site on the same PHD finger, or by using tandem PHD fingers, like in DPF3b. The set of currently characterized domains suggests that different PHD fingers can read specific combinations of post-translational marks. For example, BPTF and similar PHD fingers will read an H3 tail that is unmodified at R2 and di- or tri-methylated at K4 (Figure 4A). Whereas AIRE reads completely unmodified H3 tails (i.e. R2me0 + K4me0), BHC80 and DNMT3L read an H3 tail containing an unmodified K4, and are largely indifferent to the R2 methylation state (Figure 4B). PYGO is an example of a PHD finger that requires K4 to be methylated (with a slight preference for K4me2, at least when complexed with HD1), but is indifferent to the methylation state of R2 (Figure 4D), whereas RAG2 seems to prefer a ligand with both R2 and K4 in a methylated state (Figure 4C). The most extreme case of combinatorial reading is the tandem PHD finger in DPF3b, which requires H3K14 to be acetylated, and both H3K4 and H3R2 to be unmodified. This example provides a nice illustration of how sets of domains can be used to read more complex combinations of histone modifications, thus reinforcing the notion that distinct patterns of histone modifications are used to regulate chromatin-based gene transcription through molecular interactions with multi-protein complexes of chromatin regulators and transcription factors.
Concluding remarks
All reported PHD finger structures bind histone H3 through interactions with the first six N-terminal residues of H3, with the exception of DPF3b-PHD1 which recognizes H3K14ac. Despite the structural variations and the differences in ligand binding specificity, unifying features manifested in five key points are crucial for PHD finger–histone H3 recognition. These points are best illustrated in the PHD finger of BPTF or BHC80 when bound to an H3 peptide in a different state: (i) a histone H3 ligand always adopts an anti-parallel β-strand forming an extension of the two-stranded β-sheet of the PHD finger. The N-terminal amine of H3 form hydrogen bonds with backbone atoms of protein residues in the loop connecting to the C-terminal α-helix (loop 2); (ii) a methylated H3K4 is typically recognized by an aromatic cage formed on one surface of the central β-sheet. A change in the cage residues from aromatic to acidic amino acids alters the ligand binding preference to unmodified H3K4; (iii) recognition of H3R2 at a binding pocket that shares a common residue with that for H3K4 binding influences the overall histone state reading capacity of a PHD finger; (iv) histone state reading is further enhanced by cooperative capacity gained through tethering two or more PHD fingers in tandem. Note that many chromatin- and transcription-associated proteins contain more than two PHD fingers in tandem in a single protein (e.g. four are present in tandem in the NSD1 lysine methyltransferase). Understanding of functional cooperativity of such PHD fingers awaits further study. Finally, (v) whether a histone modification-directed interaction with a PHD finger protein results in gene activation or suppression is not dictated by the particular histone modification, but rather is dependent upon the functional context in which a given interaction takes place.
In comparison with other histone binding modules such as bromodomains and chromodomains, it is striking to note that PHD fingers - the smallest in size - are capable of exerting such complex and sophisticated functional versatility as epigenome readers. The structure-function relationships of the PHD fingers revealed from these recent studies illustrate how functional diversity of a protein module can be achieved by evolutionary changes to the structural features or amino acid residues near ligand binding sites outside the conserved scaffold. Because of their low sequence conservation and adaptable structural plasticity, it will not be surprising to see other unexpected modes and/or cooperativity of ligand recognition emerging from PHD fingers in interactions with their biological ligands, particularly histones.
Acknowledgement
The work in the authors’ laboratories was supported by grants from the National Institutes of Health (to M.-M.Z. and R.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Strahl B, Allis C. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis C. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Taverna SD, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruthenburg AJ, et al. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8(12):983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12(5):659–665. [PMC free article] [PubMed] [Google Scholar]
- 7.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 9.Maurer-Stroh S, et al. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28(2):69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen A, et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18(22):6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs S, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295(5562):2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 12.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116(2):259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33(4):428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vezzoli A, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17(5):617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 15.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 16.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31(1):35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 19.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442(7098):96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taverna SD, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24(5):785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura Y, et al. Crystal structure analysis of the PHD domain of the transcription co-activator Pygopus. J Mol Biol. 2007;370(1):80–92. doi: 10.1016/j.jmb.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1111. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange M, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22(17):2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L, et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466(7303):258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan A, et al. Engineering a protein scaffold from a PHD finger. Structure. 2003;11(7):803–813. doi: 10.1016/s0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung T, et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33(2):248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Ingen H, et al. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure. 2008;16(8):1245–1256. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Fiedler M, et al. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30(4):507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramon-Maiques S, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104(48):18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen H, et al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem. 2010;285(13):9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty S, Zeng L, Zhou MM. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure. 2009;17(5):670–679. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9(4):370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ooi S, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448(7154):718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chignola F, et al. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 2009;37(9):2951–2961. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282(4):2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17(15):1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416(6876):103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, et al. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15(1):109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28(4):677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He F, et al. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010;18(9):1127–1139. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Tsai WW, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141(7):1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]