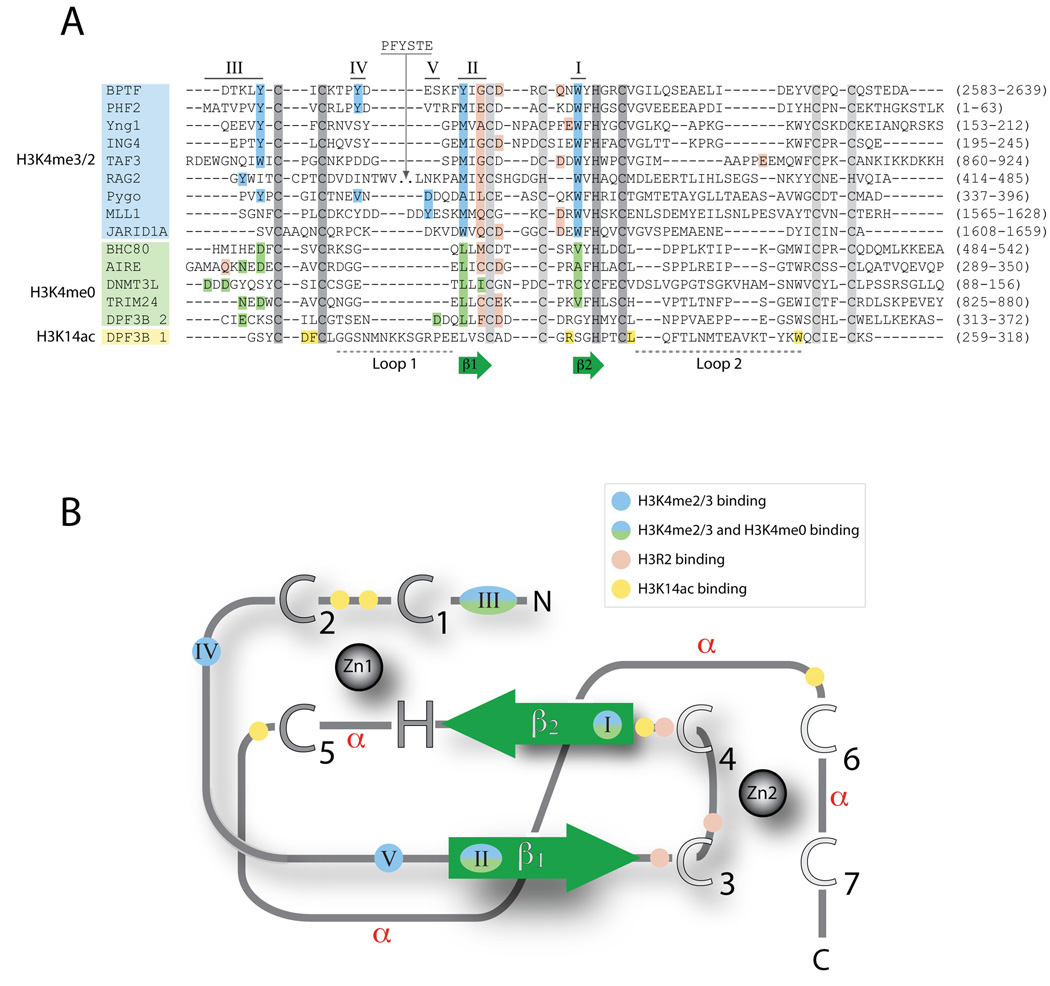
Figure 1. Conserved features of PHD fingers.
PHD fingers exhibit high sequence variability, but share some common sequence and structure patterns. Common features include the Zinc-coordinating residues, two core β-strands (β1 and β2), and the use of common sequence regions to form ligand binding sites. (A) Structure-based alignments of representative PHD finger sequences. The conserved Zinc-coordinating residues are shown in dark gray for Zinc 1 and light gray for Zinc 2, and the two core β-strands are shown in green. The regions involved in ligand recognition are labeled I through V, ordered by the frequency with which they are utilized within the domain family. Residues are colored according to their participation in the recognition of H3 tail residues: blue, H3K4me3 recognition; green, H3K4me0 recognition; pink, H3R2 recognition; and yellow, H3K14ac recognition. The residue numbers corresponding to the PHD finger in the full-length protein are shown in parenthesis. (B) Schematic of the PHD fold. All PHD fingers adopt the same basic topology. Zinc atoms are shown as gray spheres, with Zinc-coordinating residues indicated as large uppercase letters. The two core β-strands are shown in green. The regions involved in ligand recognition are labeled (I–V) and colored as indicated in A. Regions that adopt an α-helical conformation in some PHD fingers are indicated (α).