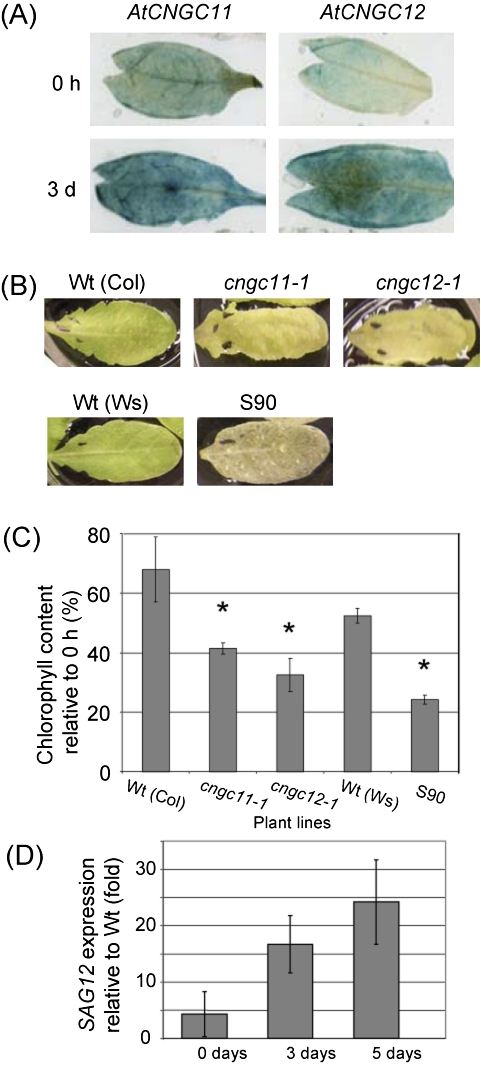
Fig. 5.
cngc11-1, cngc12-1, and S90 show altered senescence phenotypes. All experiments were repeated >3 times with similar results. (A) Expression analysis of AtCNGC11 and 12 using detached leaves from promoter:GUS transgenic plants. The leaves were floated on water in the dark for 0 d or 3 d. (B) cngc11-1 and cngc12-1, as well as S90, showed a more rapid loss of chlorophyll when detached leaves were placed in darkness for 4 d. (C) Quantitative analysis of chlorophyll degradation after 5 d in darkness. Asterisks indicate statistically significant differences (Student's t-test, P <0.05) (D) SAG12 expression levels were measured by quantitative RT-PCR in detached S90 leaves after 0, 3, and 5 d in darkness and graphed relative to SAG12 expression in Ws wild-type controls. The data shown here are the mean of three technical repeats using one cDNA sample. The same experiment was conducted three times using different cDNAs (biological repeats). The data shown here are one representative.