Figure 1.
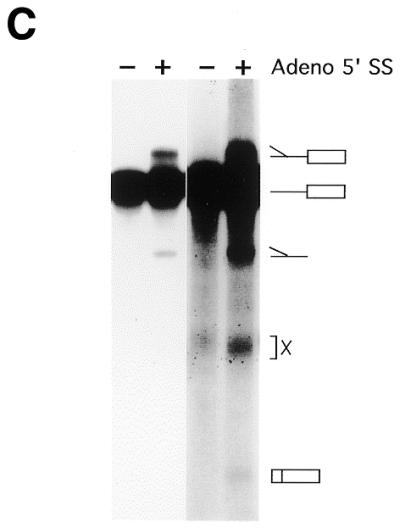
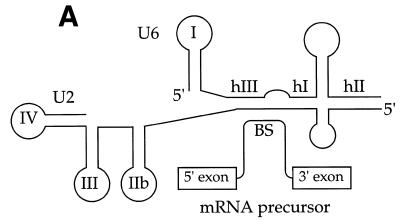
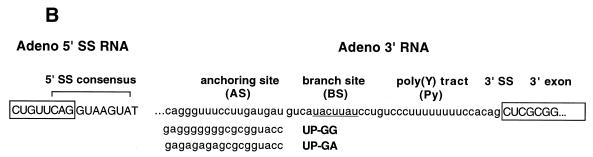
Substrates, crosslinks and trans-splicing assay. (A) U2 snRNA interactions with U6 snRNA and the mRNA precursor. U snRNA stem–loops are numbered conventionally (4); U2/U6 helices I, II and III are indicated as hI, hII and hIII; the branch site is denoted BS; and the exons are boxed. Partial, complete or alternative base pairing is denoted by parallel lines, loops and bulges by curved lines. U2 snRNA can in principle pair with the BS or with U6 to extend helix III. Adapted from (88). (B) trans-splicing substrates. The Adeno 5′ SS is a synthetic oligonucleotide with a 3′ terminal deoxythymidine to prevent degradation. The Adeno 3′ RNA contains the anchoring site (AS), the most probable BS (underlined; see 24, fig. 3), polypyrimidine tract (Py), 3′ splice site (3′ SS) and 3′ exon. (C) In vitro trans-splicing. 32P-labeled Adeno 3′ RNA was incubated with unlabeled Adeno 5′ SS oligonucleotide in HeLa nuclear extract at 30°C for 120 min. RNA was purified and resolved by denaturing 12% PAGE. The positions of Y-branched splicing intermediate, Adeno 3′ RNA, Y-branched intron and X are shown on the right. X is an exonuclease degradation product of Adeno 3′ RNA resulting from protection in the BS region (89–93). Left to right, lanes 1–4. Splicing intermediates are seen most easily on short exposure (lanes 1 and 2), ligated exons on long exposure (lanes 3 and 4).